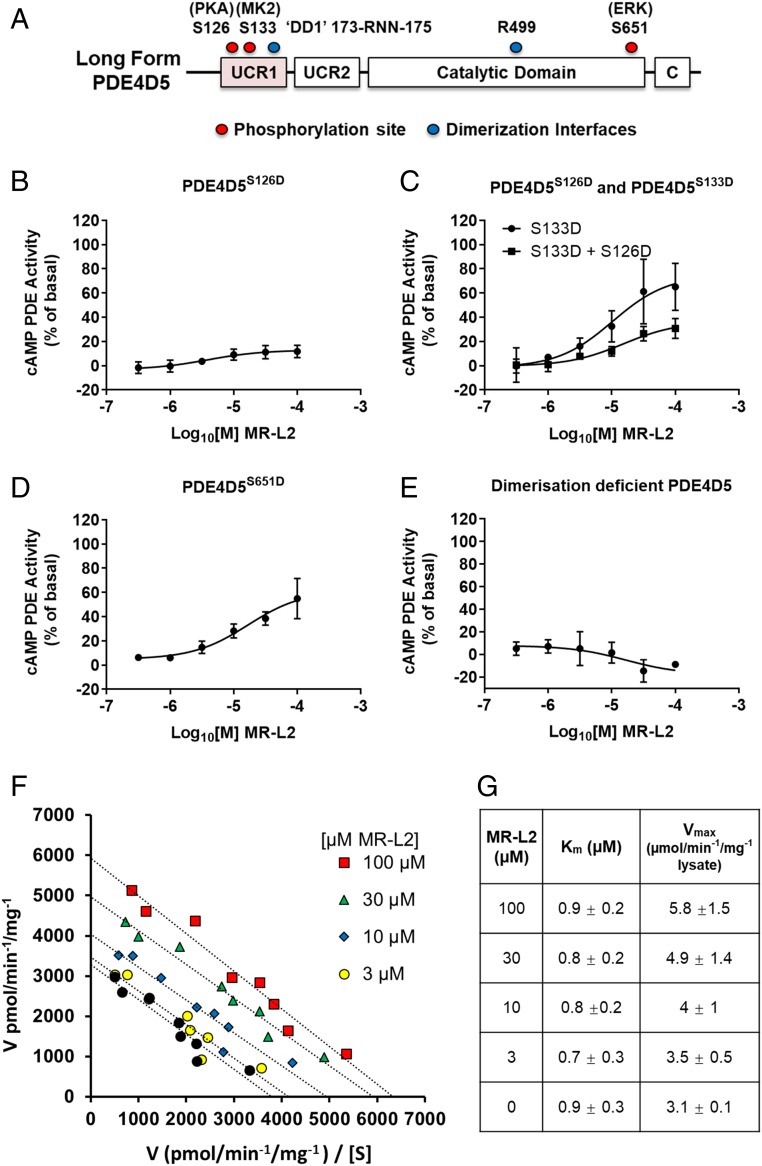
Fig. 2.
PDE4 activator treatment phenocopies PKA-mediated activation of long PDE4 isoforms. (A) A schematic diagram of PDE4 that details the mutations used to investigate the action of MR-L2 on the long PDE4 isoform, PDE4D5. (B) A S126D phosphomimetic substitution was used as a surrogate for PKA phosphorylation. The S126D mimetic construct is not activated by MR-L2, indicating that the activated long-form PDE4 state engendered by the mutation is insensitive to further pharmacological activation. (C) S133D was engineered as a surrogate for MKII phosphorylation of PDE4D5. Alone, the S133D substitution does not affect the activation of PDE4D5 by MR-L2. However, when engineered in combination with the PKA phosphomimetic, the S133D partially restores the sensitivity to pharmacological activation of the A S126D mutant enzyme. (D) The phosphorylation of PDE4D5 by Erk was similarly simulated by introducing a S651D mutation. MR-L2 activated this construct, indicating that the activator can overcome Erk phosphorylation-mediated inhibition of long-form PDE4 activity. (E) A dimerization-deficient mutant of PDE4D5 (DD1-R499D-PDE4D5) is unable to be activated by MR-L2. (F) Eadie Hofstee analysis of enzyme velocity showing the pharmacological activation of PDE4D5 with 3, 10, 30, and 100 µM MR-L2. The activator compound increases PDE4D5 Vmax without changing Km for cAMP. (G) Average Vmax and Km values calculated from n = 4 independent experiments ± SD.