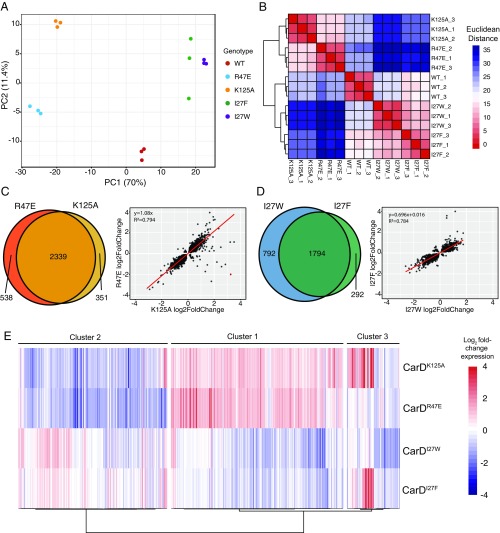
Fig. 3.
CarD mutants with similar effects on RPo stability show similar gene expression profiles. (A) A principal component analysis of RNA sequencing samples based on read counts generated by mapping reads to a library of protein encoding sequences from the Mtb H37Rv genome. Each point represents one sequencing sample colored by genotype, and the distance between 2 points reflects the variance in gene expression between the 2 samples. The first 2 principle components, PC1 and PC2, define the x- and y-axes, respectively, and account for 70% and 11.4% of the variance, respectively. (B) Hierarchical clustering of RNAseq samples based on read count data in which each column and row represents one sample. The color of each cell represents the Euclidean distance, calculated according to relative expression of all Mtb protein encoding genes, between each sample pair. (C and D) Venn diagrams and scatter plots show the overlap in the lists of Mtb genes that were significantly differentially expressed (Padj < 0.05) between (C) CarDR47E and CarDK125A, and (D) CarDI27F and CarDI27W. Scatter plots compare gene expression changes for each pair of mutants. Each point represents an Mtb gene that was significantly differentially expressed in one or both of the mutant strains being compared. The position of the dot along the axes represents the log2 fold change relative to CarDWT, with one mutant on the x-axis and the other on the y-axis. Red lines represent a linear regression line calculated for each pair. (E) Unsupervised hierarchical clustering (Pearson distance, Ward’s method linkage) of patterns of gene expression changes in a subset of 432 Mtb genes that were significantly differentially expressed greater than 2-fold in at least one CarD mutant Mtb strain relative to CarDWT. Each column represents a different gene in the set, and each row represents a different CarD mutant strain. The coloring of each cell shows the log2 fold change in expression for each gene in a given CarD mutant strain relative to CarDWT.