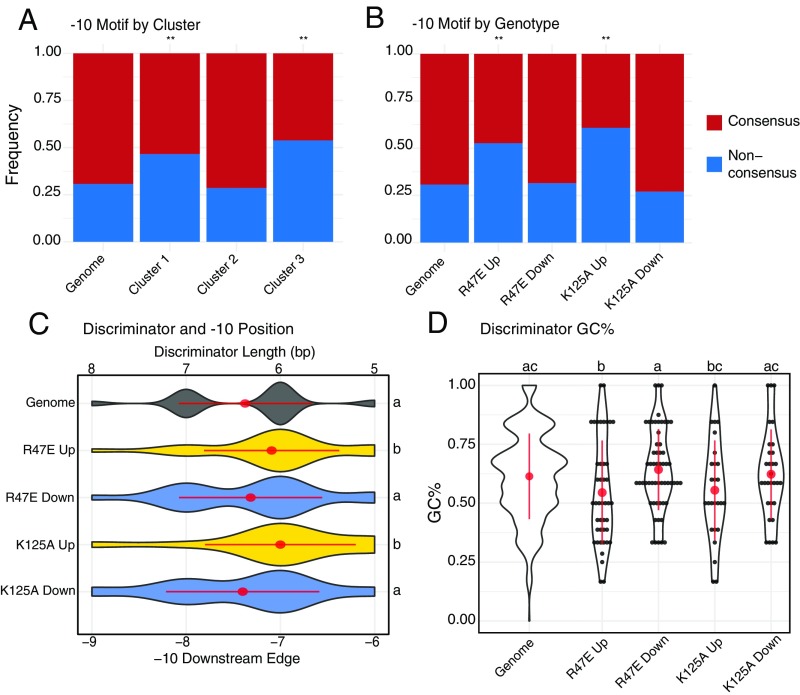
Fig. 4.
Genes that are up-regulated in RPo-destabilizing CarD mutant Mtb strains are associated with promoters lacking a consensus “TANNNT” −10 element motif and containing shorter and less GC-rich discriminator. Mtb genes directly downstream of a primary TSS identified in Cortes et al. (31) were classified on the basis of the presence of “TANNNT” DNA sequence motif in their promoter −10 element. (A and B) The relative proportions of these promoter classes are shown for the entire Mtb genome (“Genome”), (A) in gene clusters defined in Fig. 3E, and (B) in subsets of genes that were significantly up- or down-regulated in CarDR47E or CarDK125A. Nonconsensus promoters were significantly overrepresented (hypergeometric test; *P < 0.05; **P < 0.005; n.s. = not significant) in genes up-regulated in CarDR47E and CarDK125A. (C) Violin plots showing the distribution of discriminator lengths for Mtb promoter subsets. Promoters without an identifiable −10 element were not included. (D) Violin and dot plots showing distribution of discriminator GC% for Mtb promoter subsets. Red dots and lines represent mean discriminator length or GC% ± SD. Statistically significant differences in discriminator length and GC% were detected using a Kruskal-Wallis rank sum test followed by post hoc pairwise Dunn’s tests in which groups with different letters (a, b, or c) are significantly different from each other (unadjusted P < 0.05). Genome, n = 1,778; R47E Up, n = 55; R47E Down, n = 73; K125A Up, n = 46; K125A Down, n = 37; cluster 1, n = 88; cluster 2, n= 91; cluster 3, n = 26.