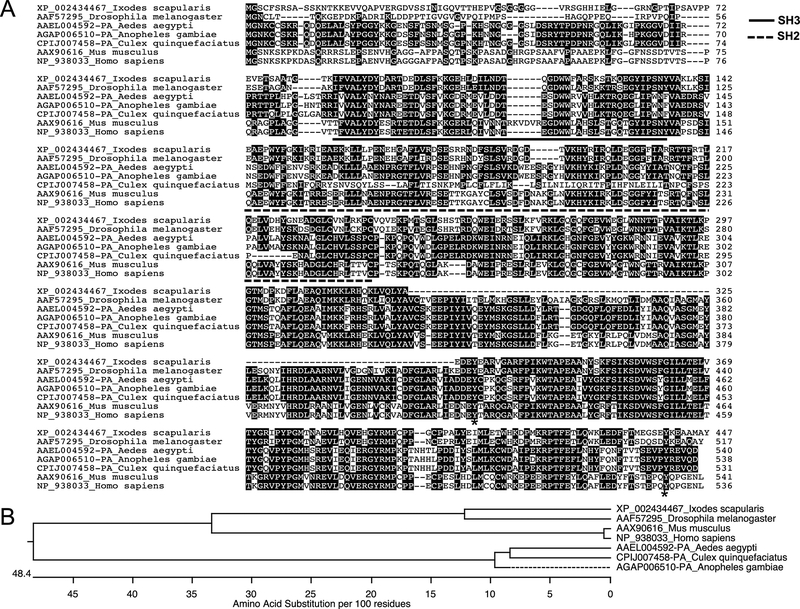
Figure 1: Alignment and phylogenetic analysis of I. scapularis Src tyrosine kinase.
A) The deduced I. scapularis Src tyrosine kinase amino acid sequence alignment (with other orthologs) using ClustalW program in DNASTAR Lasergene is shown. Residues that match are shaded in black color. GenBank accession numbers for D. melanogaster, A. aegypti, A. gambiae, C. quinquefasciatus, M. musculus, and H. sapiens Src sequences are shown. VectorBase accession numbers for I. scapularis, A. aegypti, A. gambiae and C. quinquefasciatus Src are provided. The NCBI accession numbers used for D. melanogaster, M. musculus and H. sapiens are also provided. Total length of the amino acid sequence is provided at one end (on the right) of each sequence. Asterisk in the sequence indicates possible tyrosine phosphorylation sites involved in Src activation and deactivation. B) Phylogenetic analysis was performed in DNASTAR by ClustalW slow/accurate alignment method using Gonnet as default value for protein weight matrix. Dotted line indicates variation in the A. gambiae forming a sub-clade; in comparison to the A. aegypti and C. quinquefasciatus Src. Scale at the bottom denotes amino acid substitutions per 100 amino acid residues.