Abstract
Characterization of single cell metabolism is imperative for understanding subcellular functional and biochemical changes associated with healthy tissue development and the progression of numerous diseases. However, single-cell analysis often requires the use of fluorescent tags and cell lysis followed by genomic profiling to identify the cellular heterogeneity. Identifying individual cells in a non-invasive and label-free manner is crucial for the detection of energy metabolism which will discriminate cell types and most importantly critical for maintaining cell viability for further analysis. Here, we have developed a robust assay using the droplet microfluidic technology together with the phasor approach to Fluorescence Lifetime Imaging Microscopy (FLIM) to study cell heterogeneity within and among the leukemia cell lines (K-562 and Jurkat). We have extended these techniques to characterize metabolic differences between proliferating and quiescent cells—a critical step toward label-free single cancer cell dormancy research. The result suggests a droplet-based non-invasive and label-free method to distinguish individual cells based on their metabolic states, which could be used as an upstream phenotypic platform to correlate with genomic statistics.
Keywords: Metabolism, Fluorescence Lifetime Imaging Microscopy, Phasor Analysis, Droplet microfluidics, Single cell analysis, circulating tumor cells, quiescent stage
Introduction
Single-cell analysis is important to study heterogeneity among individual cells which can lead to an improvement in disease diagnosis and treatment strategies(1,2). The ability to confine single cells in individual droplets enabled microfluidic devices to evolve as a valuable tool for single cell analysis(3). In general, single-cell encapsulation in droplets is performed randomly by optimizing the concentration of cells in the sample solution such that no droplets contain more than one cell in them. In such cases, the encapsulation efficiency is as low as 5%, which is dictated by Poisson statistics. However, the deterministic single cell encapsulation involves both active and passive methods(4). Active techniques include both laser-induced optical trapping(5) and pico-ejection techniques(6). These techniques can deliver 100% single cell encapsulation efficiency; however, the throughput is very low (<1 Hz). Passive techniques utilize hydrodynamic concepts to perform high-efficiency single cell encapsulation. One of the passive techniques utilizes the Raleigh-Plateau instability jet breakup at the flow focusing junction such that the droplets with a single cell will be larger than the empty ones. The lateral induced drift caused by the unequal oil flow rates and steric interaction at the bifurcation enable droplet sorting at 79.2% efficiency(7). Another method utilizes inertial microfluidic principles to order the cells along the channel wall in a curved channel or in a long, high aspect ratio microchannel to perform single cell encapsulation at 80% efficiency(8,9). Recently, our group has reported a microvortex based technique that can perform both high-efficiency single cell encapsulation (~50%) and size-selective cell capture in a single microfluidic device at low cell loading density(10). In this study, we used Poisson-based single cell encapsulation in droplets. The cell concentration and the flow rates are optimized to achieve maximum encapsulation efficiency with no doublets or triplets.
Encapsulating single cells is merely the first step in single cell analysis. Once cells are captured, identification becomes difficult given that they no longer exhibit a phenotypic profile or significant morphological difference. Current platforms to perform single-cell studies includes flow cytometry(11), and automated microscopy(12). Examples of single cell analysis in droplets include screening single cells against the library of drug compounds(13), genome-wide expression profiling using Dropseq(14) and inDrop(15) methodologies. Notably, the single-cell analysis in droplets mostly involves cell lysis and genomic analysis including PCR and sequencing to identify the gene expression level. However, genotyping relies on end results, and fluorescent labeling or cell lysis is not desirable for continuous and dynamic single cell monitoring.
To meet the unmet need for single cell detection and identification, we have developed Fluorescence lifetime imaging microscopy (FLIM) which is a non-invasive, label-free assay that measures energy metabolism in encapsulated cells. Metabolism is fundamentally important for identification between healthy and unhealthy cells, tumor heterogeneity, wound healing, diabetes, and neurodegenerative disorders(16–19). The quantitative measurement of the metabolites can be applied as a diagnostic and drug screening tool due to the reflection of the changes during disease development and treatment response(20–22). The accurate determination of metabolites from a single cell is critical.
The pioneering work of Britton Chance has opened the field of spectroscopy and optical imaging to detect metabolic intrinsic fluorescent co-factors in cells and tissues (23,24). These spectroscopic techniques provide information of the natural biomarkers involved in different stages of the electron transport chain, such as the reduced form of nicotinamide adenine dinucleotide (NADH), nicotinamide adenine dinucleotide phosphate(NADPH) and flavins (FAD)(25,26). Skala et al. have used fluorescence intensity of FAD divided by the fluorescence intensity of NADH to characterize the redox ratio in living cells(27). FLIM has also been used to determine the changes in metabolism from fluorescence lifetime of NADH and FAD(27–30). We also apply the phasor analysis with FLIM, in which the Fourier cosine and sine transforms of the experimental decay are computed and represented by polar coordinates “g” and “s” in the transformation function on a polar coordinate graph termed the ‘phasor plot’.(31,32) Phasor approach provides fit-less and graphical data analysis which is more intuitive(31,33–35). We apply 740nm to excite the intrinsic fluorescent species which can give the maximum excitation for NAD(P)H(36). Given that endogenous molecules such as collagen, retinoids, flavins, folate, and NAD(P)H are intrinsically fluorescent in live cells(37,38), fluorescence lifetime data can have a mixture of all these species making it difficult to identify these components. Using a short pass filter to collect the emission of NAD(P)H and the phasor method, contributions from these different biochemical species are easily identified and separated. Our previous work has showed a leukemia screening platform combines FLIM and microfluidic single-cell trapping which can successfully distinguish the white blood cells and leukemia cells(39). However, it has been demonstrated cell-environment may contribute to the leukemia cell metabolism, survival rate, proliferation rate, etc.(40,41) The novel microfluidic single encapsulated oil droplet can be used to isolate and maintain the cell metabolic signature. And this design can also help with the future personal therapeutic drug introduction and FLIM screening (13). Compared to the single cell fluorescence-activated cell sorting(FACS) which has been largely used in clinic, the presented metabolic screening platform doesn’t have to use the fluorescent tag with expensive antibodies which also might cause cell damage and cell behavior alteration.
Here we have applied the phasor-FLIM approach to leukemia cells and serum-free treated fibroblasts encapsulated one-to-one in droplets and have captured detailed data on their metabolic states. The encapsulated leukemia cells display heterogeneous phasor-FLIM signatures due to different proliferation rates. However, the difference between the metabolic lifetimes of the two leukemia cell types used, are still distinguishable. Furthermore, we found that serum-starved fibroblasts, under a quiescent state, deviate significantly from that of the fibroblasts cultured under normal condition, indicating that lifetime trajectories can be used to detect metabolic alterations during different stages of cell cycle. Our results show that under serum-free treatment, there is a higher free NADH and NADPH production due to increased glycolysis through the pentose phosphate pathway which may contribute to detoxify free radicals or to synthesize fatty acids(42–44). In summary, our non-invasive phasor-FLIM approach provides an intuitive, sensitive, quantitative method to assess metabolic response in encapsulated single cell droplets which can be applied for fast screening of the cancer cells and drug development.
Results and Discussion
Single Cell Encapsulation in Droplets
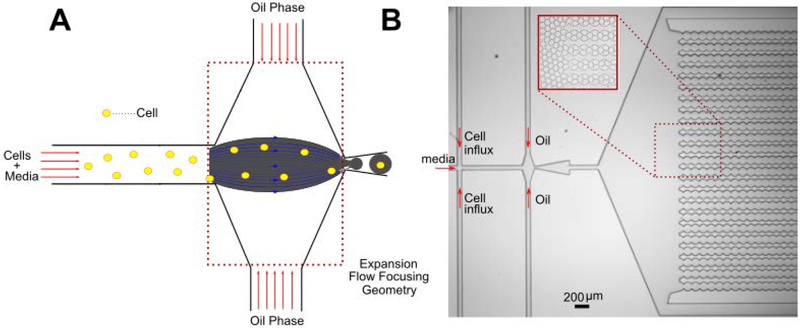
The microfluidic device has an expansion flow focusing geometry as shown in Fig. 1A. This design has been used by our group for droplet-based applications since 2006 (45). The device consists of two inlets: Cells suspended in the culture media is injected through the left inlet while the oil phase enters the device through the other inlet. Both the inlets have filters (20 μm) to prevent the debris from clogging the flow-focusing junction. The microfluidic channel height is made comparable to the cell diameter such that no two cells roll over each other. Figure 1B illustrates the microfluidic device made from Polydimethylsiloxane (PDMS). It has a unique droplet collection chamber design to prevent the droplet motion throughout the data acquisition process for FLIM.
Figure 1:
Single cell encapsulation in droplets: (A) Device schematic: Cells introduced from the left inlet are encapsulated 1–1 into the droplets which is dictated by the Poisson statistics (B) Microfluidic device made of PDMS, which is used for single cell encapsulation as well as FLIM characterization. Inset shows the droplets if collected in the uniquely designed collection chamber. It prevents the droplet motion during FLIM measurements to a considerable extent.
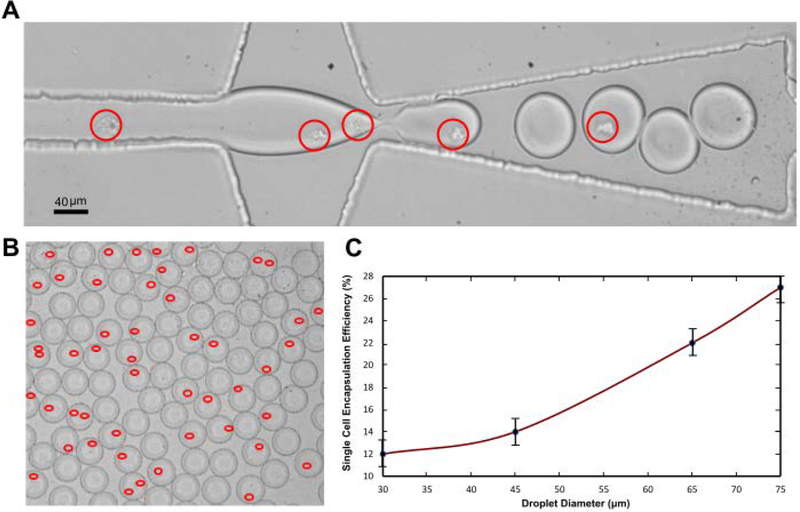
The process of encapsulation of cells one-to-one into the droplets is illustrated in Fig. 2A. The droplet diameter and droplet generation frequency is adjusted according to the cell influx to ensure that maximum encapsulation efficiency is achieved. This is achieved by precisely tuning the aqueous phase to the oil phase pressure ratio (Pa/Po). At a cell concentration of 2 × 106 per ml, we achieved a maximum encapsulation efficiency of 27 %, with a cell doublet rate of 8 % (Fig 2 C). There is a trade-off between the cell doublet rate and single cell encapsulation efficiency. If no droplets contain more than one cell (no doublets), then the single cell encapsulation efficiency decreases to <10 %. The critical parameters that influence the 1–1 efficiency are the droplet diameter, droplet generation frequency, cell concentration, and difference in cell-media density.
Figure 2:
(A) Demonstration of the single cell encapsulation process. The red circle indicates the presence of one cell. (B) Droplets collected after the encapsulation process. The presence of cells is highlighted in red. (C) Graphic illustrating the encapsulation efficiency of k-562 cells in droplets. Efficiency increases with the droplet diameter and peaks at an optimum diameter. However, it is observed that the efficiency decreases beyond a threshold diameter due to multiple encapsulations (doublets/triplets) (not shown).
Oil Phase Characterization for best FLIM performance
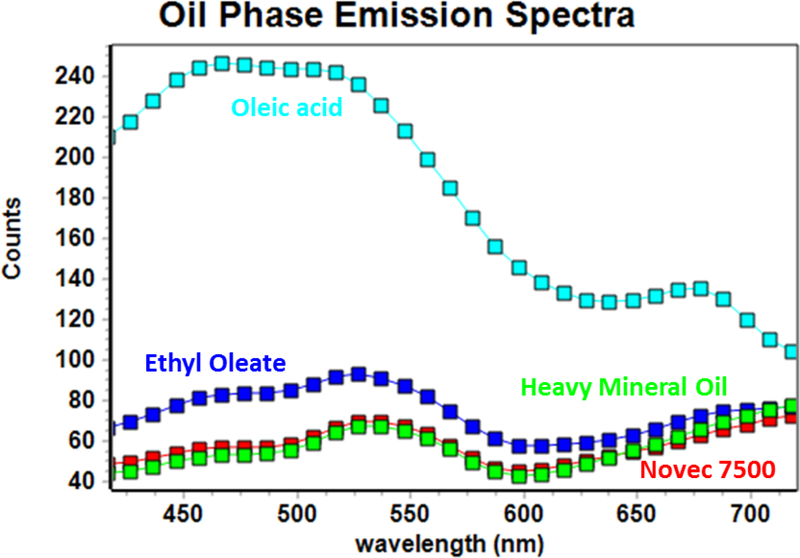
In order to ensure the FLIM performance, we determined the optimum oil phase to avoid background signal from oil phase, while allowing the rapid and robust acquisition of the FLIM signal on cells encapsulated in the droplets. We screened four different oil phase solution, Novec 7500 mixed with Dolomite picosurf (mix in the ratio of 4:1), ethyl oleate and Ebil EM90, heavy mineral oil with 2% Ebil EM90 and 0.1% Triton X-100 as well as oleic acid with 2% Span 80. To quantify the fluorescence intensity and emission spectra from the oil phase, we expose the 740nm 2-photon laser to the solutions. The emission spectra (Fig. 3) shows that Novec 7500 mixed with Dolomite picosurf (mix in the ratio of 4:1) and heavy mineral oil with 2% Ebil EM90 and 0.1% Triton X-100 present lowest fluorescence intensity which will not interfere with the FLIM imaging collection. Novec 7500 has been used in pharmaceutical and chemical manufacturing processes since its property of cell-friendly and stability which is optimal for live imaging and the down-stream PCR test in the future. Therefore, we choose Novec 7500 as our oil phase for the high-efficiency one-one droplet encapsulation platform.
Figure 3.
Oil Phase Emission Spectra test(740nm excitation) for Oil 1(red)- Novec 7500 mixed with Dolomite picosurf (mix in the ratio of 4:1), Oil 2(blue)- ethyl oleate and Ebil EM90, Oil 3(green)- heavy mineral oil with 2% Ebil EM90 and 0.1% Triton X-100, Oil 4(cyan)- oleic acid with 2% Span 80.
Differentiation of Leukemia Cell lines in Droplets using FLIM
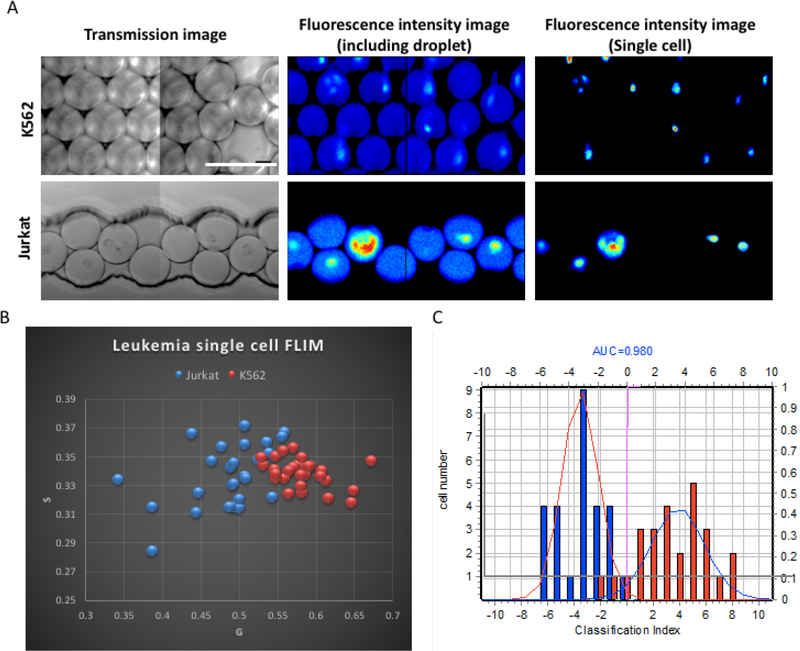
Two different types of human leukemia cells K562 erythromyeloid and Jurkat T-cell leukemia were encapsulated one-to-one in the droplets (Fig. 4). FLIM images were collected at 740 nm excitation wavelength. The transmission images of the cells in the device, the fluorescence intensity images of the single cells with and without droplet are shown in Figure 4A and 4B. We distinguish the different leukemia cell lines by morphology and phasor-FLIM signature (Supplementary Fig. 1, Fig. 4C, D).
Figure 4.
A) Transmission images, fluorescence intensity images of the K562 and Jurkat cells respectively within droplets and without droplets. Scale bar: 100 μm. B) Scatter plot shows the zoomed in phasor-FLIM results of the single leukemia cells in the high efficiency encapsulated droplets for both K562 (red) and Jurkat (blue) cells. The different leukemia cell lines K562 and Jurkat have significant difference, with a p-value for g, 0.025 and s, 0.0002. C) Bar graph of classification index of leukemia cell lines. K562 cells are shown in red, Jurkat cells are in blue. The area under curve (AUC) is 0.980. (K562 group, n=33, Jurkat group, n=32).
Most of the normal leukemia cells have a diameter ranging from 8μm to 20μm, and there exist significant size overlap based on our measurement of the leukemia cell lines we investigated, K562 and Jurkat cells (Supplementary Fig.1). The diameter of the K562 and Jurkat cells are 11.15 μm and 10.48 μm, respectively, which shows no significant difference and cannot be distinguished based on morphology. While leukemia cells cannot be differentiated by size and shape, the FLIM phasor shows significant difference without any label. We applied phasor transformation to the acquired FLIM data and plotted the representative scatter plot of phasor-FLIM signature of the leukemia cells in droplets (Fig. 4C), the Jurkat cells (blue) and K562 cells (red) show a significant difference.
To further quantify the difference of the phasor-FLIM signatures between different cell populations and the cellular heterogeneity within the same population, we apply Distance analysis to plot the 3D phasor information of individual cells of Jurkat (blue), and K562 (red) in the bar graph of Fig. 4D(46). Both of K562 erythromyeloid cells and Jurkat T-cell leukemia cells demonstrate a relatively shorter lifetime compare to normal cells and tissues(34), and as shown, the index shows the cell phasors of the Jurkat cells are significantly different from the group of K562 cells. These can be explained by the Warburg Effect, in which rapid-proliferating tumor-like cells, i.e., leukemia cells, have stronger glycolysis in glucose metabolism to support fast ATP consumption and biosynthesis of macromolecules and there is tumor heterogeneity due to different metabolic rate.
Phasor-FLIM measurement of regular and quiescent fibroblast cells in droplets
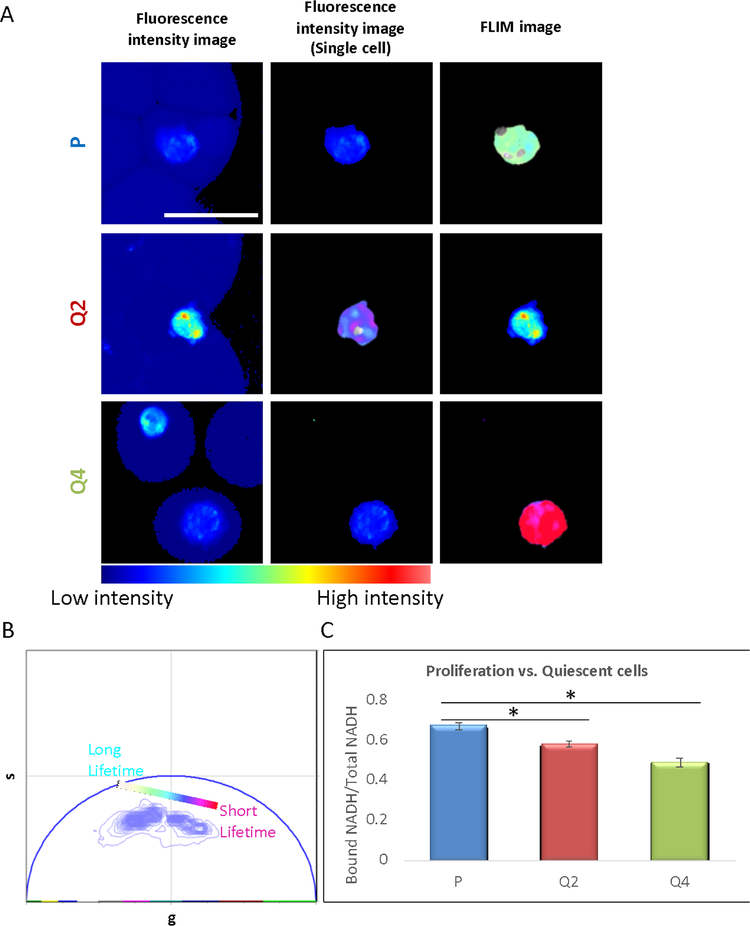
After identification of the difference in the phasor-FLIM fingerprints of different leukemia cell lines (Jurkat and K562), we conducted the rapid label-free single-cell screening to understand the quiescent and proliferation metabolic activity and to further observe the rapid treatment response. The model we used to monitor the metabolic difference between cellular quiescence is human foreskin fibroblast model. Cells from human foreskin fibroblasts cell line were treated for 24 hours(Q2) and 72 hours(Q4) with non-serum media, and the phasor-FLIM signature of each condition was measured(Fig. 5). We collected the NAD(P)H fluorescence emission of encapsulated single cells and fluorescence lifetime images at 740nm excitation and pseudo-colored the higher bound/free-NAD(P)H-ratio group as cyan and the higher free/bound-NAD(P)H-ratio group as pink (Fig. 5A, B). The human foreskin fibroblasts with different treatment type of serum starvation were clearly distinguished from the cells treated using normal condition as highlighted in phasor plot (Fig. 5B) as serum-starved human foreskin fibroblast cells have more components in the color indicating a higher fraction of free NAD(P)H, while normal human foreskin fibroblasts consist of more white color pixels indicating a higher fraction of bound NAD(P)H. The fraction of bound NAD(P)H/total NAD(P)H shows the quantification of significant increase of the free-form of NAD(P)H with serum-free treatment or the induction of cells in a quiescent state. The 72 hours serum-starvation (Q4) group has a significant shift from high NADPH bound fraction to a lower bound fraction (Fig. 5c). This shift, toward a higher free fraction of NADPH and shorter lifetime, in the quiescent group (Q4) compared to the proliferating group (P) may be caused by the metabolic shift towards the Warburg effect (increased glycolysis in the presence of oxygen) and the pentose phosphate pathway (PPP) pathway (42,47). Under this glycolytic state, introduced by serum-starvation, there would be a higher lactate production that can create an acidic microenvironment for gene expressions regulation (48–50). This is important in tumor progression and wound healing promotion. In addition, the PPP pathway can produce more ribose-5-phosphate and NADPH which is needed for the biosynthesis of nucleotides and macromolecules including fatty acids. Given that previous reports have correlated a link between human newborn fibroblasts and cancer cells in regards to similar migratory properties, the screening between cellular state might open a new window of further understanding of the basic mechanisms involved in communication between tumor cells and fibroblasts as well as cancer dormancy for targeted cancer therapy(51).
Figure 5.
A) Fluorescence intensity images of the single cell within droplets, and FLIM images of the human foreskin fibroblast under regular proliferation condition(P), serum starved for 2 days(Q2), and serum starved for 4 days(Q4). The FLIM images were pseudocolored by the color bar in Fig. 5B, scale bar: 50 μm. B) Phasor plot shows the FLIM signature distribution of the P, Q2 and Q4 conditions. From Cyan to Pink, the color shows the signature move from the long lifetime (higher Bound/Free NAD(P)H) to the short lifetime (lower Bound/Free NAD(P)H). C) Bar graph shows the bound NAD(P)H/Total NAD(P)H ratio of the P, Q2 and Q4 conditions. The Q2 and Q4 group shows significant decreasing of Bound NAD(P)H, p-value for P and Q2 groups is 0.001, p-value for P and Q4 group is 0.002 (P group, n=21, Q2 group, n=4, Q4 group, n=19).
Materials and Methods
Microfluidic Device Preparation.
Microfluidic devices were fabricated in polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) using soft lithography(52). The PDMS molded imprints and the plain glass slide is plasma treated (Harrick Plasma Inc) for 2 minutes and were brought together to form a permanent seal. The device is left in the oven at 120 ° C overnight to regain its natural hydrophobicity.
Fluidic setup, Leukemia cell preparation and Imaging.
In the experiments,Novec 7500 mixed with Picosurf surfactant (5 % w/w) (Dolomite) constitute the continuous phase, and the cells suspended in the culture media forms the dispersed phase. Both the K-562 cells and Jurkat cells (American Type Culture Collection (ATCC®)) were cultured in a T-75 cell culture flask using RPMI 1640 as a basal medium to which fetal bovine serum (FBS) was added (10% by volume). The cell culture media is changed every three days until the cells proliferate to the desired confluency. The cells containing medium were then transferred to a 10 mL Eppendorf tube, centrifuged at 1000 rpm for 5 min and the pellet was re-suspended in the freshly prepared aqueous/dispersed phase. The cell concentration in the resulting suspension was determined using an automated cell counter (ThermoFisher Scientific) and appropriately diluted to the desired concentration.
We used constant pressure source via high-speed solenoid valves controlled by a custom-built lab view program to automate the fluid injection into the microfluidic chip. The single cell encapsulation process was monitored using Nikon 100-S inverted microscope and recorded using Phantom camera, V-310 (Vision Research). To analyze the videos frame by frame to yield the encapsulation data, we used ImageJ, a public domain java based image processing software developed at National Institutes of Health(53).
Fluorescence Lifetime Imaging Microscopy.
Fluorescence lifetime images of the cells encapsulated in high-efficiency microdevice were acquired on Zeiss LSM880 (Carl Zeiss, Jena, Germany), a multi-photon microscope coupled with a Ti:Sapphire laser (Spectra-Physics Mai Tai, Mountain View, CA) with 80 MHz repetition rate. The FLIM data detection was performed by the photomultiplier tube (H7422p-40, Hamamatsu, Japan) and a320 FastFLIM FLIMbox (ISS, Champaign, IL). The cells encapsulated in droplets were excited at 740nm. A Zeiss EC Plan-Neofluar 20x/0.8 NA objective (Cart Zeiss, Jena, Germany) was used. The following settings were used for the measurement: image size of 256×256 pixels, with a scan speed of 16μs/pixel. A dichroic filter at 690nm was used to separate the fluorescence signal from the laser light. And the emission signal is split with 496nm LP filter and detected in two channels using a bandpass filter 460/80 and a 540/50 filter. Only the blue channel (460/80) data was used for this study. FLIM calibration of the system was performed by measuring the known lifetime of a fluorophore coumarin 6 which has a known fluorescence lifetime of τ=2.5 ns when dissolved in ethanol. FLIM data were acquired and processed by the SimFCS software developed at the Laboratory of Fluorescence Dynamics (LFD).
Human Foreskin Fibroblast cells preparation.
Human foreskin fibroblast cells were cultured and maintained in a 37°C incubator humidified at 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium/F12(D-MEM) (1:1) (11320–033, Life Technologies, Carlsbad, CA), supplemented with 10% fetal bovine serum and 1% penicillin streptomycin. The cells were freshly trypsinized and plated onto 35 mm glass bottom dishes (Mattek Corporation, Ashland, Massachusetts) for imaging. The phasor-FLIM signature of NADH bound to lactate dehydrogenase and NADH and NADPH (free form) are shown in Supplementary figure 2.
Serum Starvation treatment.
Dulbecco’s modified Eagle’s medium/F12(D-MEM) (1:1) (11320–033, Life Technologies, Carlsbad, CA), supplemented with 0.1% fetal bovine serum and 1% penicillin streptomycin were prepared for the serum starvation treatment. The Q2 and Q4 cells were serum starved for 24 hours and 72 hours respectively.
Statistical analysis.
Data are presented as mean ± standard deviation. For the FLIM data, the statistical analyses were performed using student t-test for the g value and the value of the ratio of bound NAD(P)H to total NAD(P)H, p<0.05 was considered as statistically significant.
Conclusion
In summary, here we present a non-invasive, label-free single cell analysis platform combining the droplet microfluidic technology and the Fluorescence Life Time Imaging Microscopy. The single cell encapsulation technique is passive, controlled only by flow rates of the two phases and not requiring complex structures or on-chip active devices, and the label-free phasor-FLIM characterization is non-invasive without micro-environment influence. To identify between encapsulated leukemia cell types (Jurkat and K562 cells) we used the phasor-FLIM analysis and showed that excellent bi-classification (AUC: 0.980) in calculating the degree of separation between the two cell types. In another example, the differences between proliferating versus quiescence states also showed statistically significant separation in the phasor-FLIM plots. Given that cells can exhibit a quiescence state where they can exit the cell cycle by reducing their size, DNA syntheses and different metabolic activity, it would seem feasible to distinguish them from their metabolic state. Indeed our phasor-FLIM analysis showed that the fingerprint of the proliferating versus quiescent human fibroblasts under exhibit higher free NADPH under quiescent state versus proliferative state. This may be due to the production of citrate from the conversion of α-ketoglutarate through NADPH shuttling through the mitochondria. In quiescence, NADPH is generated as a means to utilize this co-factor to synthesize fatty acids. This would indicate a higher production of NADPH would be produced to detoxify free radicals or to synthesize fatty acids under a quiescent state. The phasor analysis proved to be effective in identifying between a proliferative versus a quiescent state that can be coupled to our single cell encapsulation design(42). The future directions of this project would be to further improve and optimize the parameters to maximize the encapsulation efficiency, perform large-scale FLIM characterization, and to incorporate the genotyping unit downstream to correlate with the phenotypic signature. Our finding suggests that FLIM-single oil droplet technique is promising as it expands the capabilities of droplet microfluidics to address applications related to single-cell high throughput early stage tumor and leukemia screening and personalized therapy by targeting quiescent tumor cells.
Supplementary Material
Acknowledgments:
The authors would like to acknowledge support from the National Science Foundation and the industrial members of the Center for Advanced Design and Manufacturing of Integrated Microfluidics (NSF I/UCRC award number IIP-1362165) (N.M, G.K. A.P.L. and M.A.D.). This work was also supported by the NIH grants 2P41GM103540 (M.A.D.) and the Samueli Career Development Chair (M.A.D).
Footnotes
Competing Interests: The authors declare no competing interests.
Reference
- 1.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Carlo D, Tse HTK, Gossett DR. Introduction: why analyze single cells? Single-Cell Analysis: Springer; 2012. p 1–10. [DOI] [PubMed] [Google Scholar]
- 3.Joensson HN, Andersson Svahn H. Droplet Microfluidics—A Tool for Single-Cell Analysis. Angewandte Chemie International Edition 2012;51:12176–12192. [DOI] [PubMed] [Google Scholar]
- 4.Collins DJ, Neild A, Liu A-Q, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab on a Chip 2015;15:3439–3459. [DOI] [PubMed] [Google Scholar]
- 5.He M, Edgar JS, Jeffries GD, Lorenz RM, Shelby JP, Chiu DT. Selective encapsulation of single cells and subcellular organelles into picoliter-and femtoliter-volume droplets. Analytical chemistry 2005;77:1539–1544. [DOI] [PubMed] [Google Scholar]
- 6.Schoendube J, Wright D, Zengerle R, Koltay P. Single-cell printing based on impedance detection. Biomicrofluidics 2015;9:014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabert M, Viovy J-L. Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proceedings of the National Academy of Sciences 2008;105:3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemna EW, Schoeman RM, Wolbers F, Vermes I, Weitz DA, Van Den Berg A. High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel. Lab on a Chip 2012;12:2881–2887. [DOI] [PubMed] [Google Scholar]
- 9.Edd JF, Di Carlo D, Humphry KJ, Köster S, Irimia D, Weitz DA, Toner M. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab on a Chip 2008;8:1262–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamalakshakurup G, Lee AP. High-efficiency single cell encapsulation and size selective capture of cells in picoliter droplets based on hydrodynamic micro-vortices. Lab on a Chip 2017;17:4324–4333. [DOI] [PubMed] [Google Scholar]
- 11.Bendall SC, Simonds EF, Qiu P, El-ad DA, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nature Reviews Molecular Cell Biology 2006;7:690. [DOI] [PubMed] [Google Scholar]
- 13.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proceedings of the National Academy of Sciences 2009;106:14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Cianni G, Miccoli R, Volpe L, Lencioni C, Del Prato S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes/metabolism research and reviews 2003;19:259–270. [DOI] [PubMed] [Google Scholar]
- 17.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell 2008;134:703–707. [DOI] [PubMed] [Google Scholar]
- 18.Hunt TK, Conolly WB, Aronson SB, Goldstein P. Anaerobic metabolism and wound healing: an hypothesis for the initiation and cessation of collagen synthesis in wounds. The American Journal of Surgery 1978;135:328–332. [DOI] [PubMed] [Google Scholar]
- 19.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1α in Huntington’s disease neurodegeneration. Cell metabolism 2006;4:349–362. [DOI] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England journal of medicine 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D. A Big Bang model of human colorectal tumor growth. Nature genetics 2015;47:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White RE. High-throughput screening in drug metabolism and pharmacokinetic support of drug discovery. Annual review of pharmacology and toxicology 2000;40:133–157. [DOI] [PubMed] [Google Scholar]
- 23.Chance B, Williams G. Respiratory enzymes in oxidative phosphorylation II. Difference spectra. Journal of Biological Chemistry 1955;217:395–408. [PubMed] [Google Scholar]
- 24.Chance B, Williams G. Respiratory enzymes in oxidative phosphorylation III. The steady state. Journal of Biological Chemistry 1955;217:409–428. [PubMed] [Google Scholar]
- 25.Brodie BB, Gillette JR, La Du BN. Enzymatic metabolism of drugs and other foreign compounds. Annual review of biochemistry 1958;27:427–454. [DOI] [PubMed] [Google Scholar]
- 26.Ying W NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants & redox signaling 2008;10:179–206. [DOI] [PubMed] [Google Scholar]
- 27.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 2007;104:19494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoumi A, Yeh A, Tromberg BJ. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proceedings of the National Academy of Sciences 2002;99:11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kukreti S, Cerussi AE, Tanamai W, Hsiang D, Tromberg BJ, Gratton E. Characterization of metabolic differences between benign and malignant tumors: high-spectral-resolution diffuse optical spectroscopy. Radiology 2009;254:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimitrow E, Riemann I, Ehlers A, Koehler MJ, Norgauer J, Elsner P, König K, Kaatz M. Spectral fluorescence lifetime detection and selective melanin imaging by multiphoton laser tomography for melanoma diagnosis. Experimental dermatology 2009;18:509–515. [DOI] [PubMed] [Google Scholar]
- 31.Digman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophysical journal 2008;94:L14–L16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma N, Digman MA, Malacrida L, Gratton E. Measurements of absolute concentrations of NADH in cells using the phasor FLIM method. Biomedical optics express 2016;7:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colyer RA, Lee C, Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microscopy research and technique 2008;71:201–213. [DOI] [PubMed] [Google Scholar]
- 34.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proceedings of the National Academy of Sciences 2011;108:13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci Rep 2012;2:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang S, Heikal AA, Webb WW. Two-photon fluorescence spectroscopy and microscopy of NAD (P) H and flavoprotein. Biophysical journal 2002;82:2811–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A 2011;108:13582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta R, Alfonso-Garcia A, Cinco R, Gratton E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Sci Rep 2015;5:9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D-H, Li X, Ma N, Digman MA, Lee AP. Rapid and label-free identification of single leukemia cells from blood in a high-density microfluidic trapping array by fluorescence lifetime imaging microscopy. Lab on a Chip 2018;18:1349–1358. [DOI] [PubMed] [Google Scholar]
- 40.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. science 2009;324:1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang M, Kim SS, Lee J. Cancer cell metabolism: implications for therapeutic targets. Experimental & molecular medicine 2013;45:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemons JM, Feng X-J, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA. Quiescent fibroblasts exhibit high metabolic activity. PLoS biology 2010;8:e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sagone AL Jr, LoBuglio AF, Balcerzak SP. Alterations in hexose monophosphate shunt during lymphoblastic transformation. Cellular immunology 1974;14:443–452. [DOI] [PubMed] [Google Scholar]
- 44.Barbehenn EK, Wales RG, Lowry OH. The explanation for the blockade of glycolysis in early mouse embryos. Proc Natl Acad Sci U S A 1974;71:1056–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Y-C, Cristini V, Lee AP. Monodispersed microfluidic droplet generation by shear focusing microfluidic device. Sensors and Actuators B: Chemical 2006;114:350–356. [Google Scholar]
- 46.Ranjit S, Dvornikov A, Levi M, Furgeson S, Gratton E. Characterizing fibrosis in UUO mice model using multiparametric analysis of phasor distribution from FLIM images. Biomedical Optics Express 2016;7:3519–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golpour M, Niaki HA, Khorasani HR, Hajian A, Mehrasa R, Mostafazadeh A. Human fibroblast switches to anaerobic metabolic pathway in response to serum starvation: a mimic of warburg effect. International journal of molecular and cellular medicine 2014;3:74. [PMC free article] [PubMed] [Google Scholar]
- 48.Barar J, Omidi Y. Dysregulated pH in tumor microenvironment checkmates cancer therapy. BioImpacts: BI 2013;3:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y. Acidic extracellular microenvironment and cancer. Cancer cell international 2013;13:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philp A, Macdonald AL, Watt PW. Lactate–a signal coordinating cell and systemic function. Journal of Experimental Biology 2005;208:4561–4575. [DOI] [PubMed] [Google Scholar]
- 51.Schor SL, Schor AM, Grey A, Rushton G. Foetal and cancer patient fibroblasts produce an autocrine migration-stimulating factor not made by normal adult cells. Journal of cell science 1988;90:391–399. [DOI] [PubMed] [Google Scholar]
- 52.Xia Y George m. Whitesides. Annu. Rev. Mater. Sci 1998;28:153–84. [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods 2012;9:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.