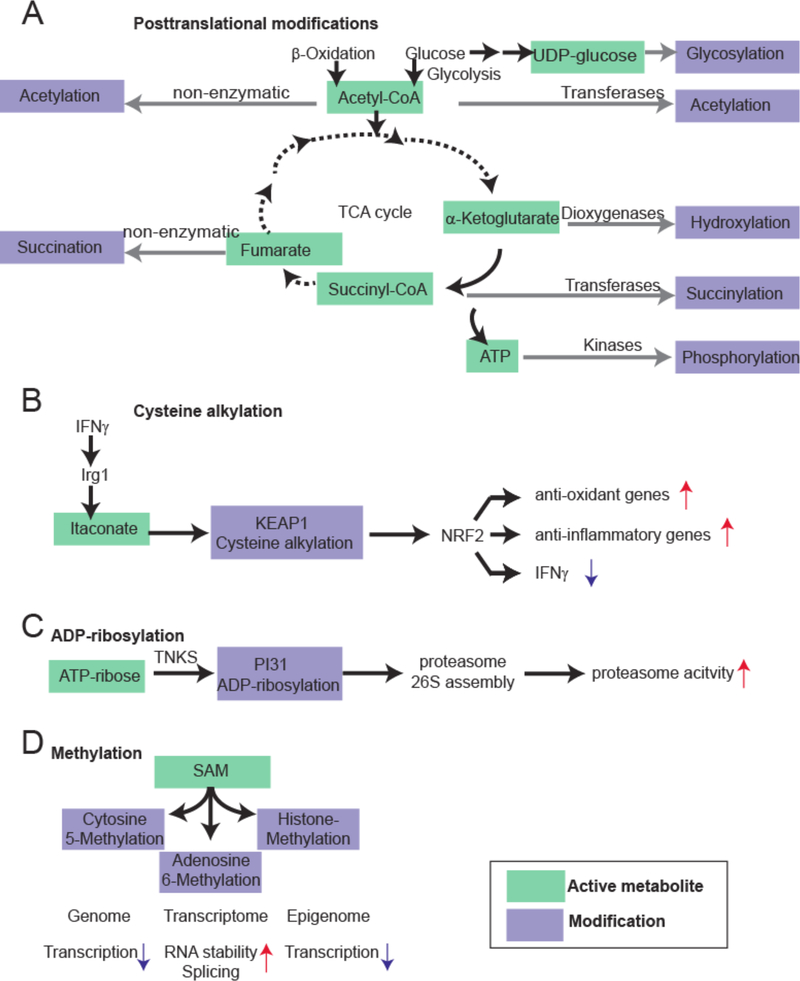
Figure 2:
Examples of macromolecule modification by the active metabolome. A. Examples for macromolecule modifications by the tricarboxylic acid cycle (TCA) intermediates and related products. Acetyl-CoA, α-ketoglutarate, Succinyl-CoA, UDP-glucose and ATP are energy-rich molecules that can directly modify proteins, or nucleic acids. B. Cysteine alkylation by itaconate, a branched fatty acid. Recently, it was shown that itaconate is an anti-inflammatory metabolite that directly alkylates cysteine residues at KEAP1 to control expression of NRF2. KEAP1 is the primary negative regulator of NRF2. This results in increased expression of anti-oxidant and anti-inflammatory gene expression. C. ADP-ribosylation of proteasome unit PI31 to control proteostasis. By the TNKS enzyme, PI31 is ADP-ribosylated to promote proteasome 26S assembly (from the 20S subunit). This results in increased proteasome activity. D. S-Adenosylmethionine is a central methyl donor for DNA, RNA and histone methylation, thereby controlling gene expression at the level of the genome, transcriptome and epigenome.