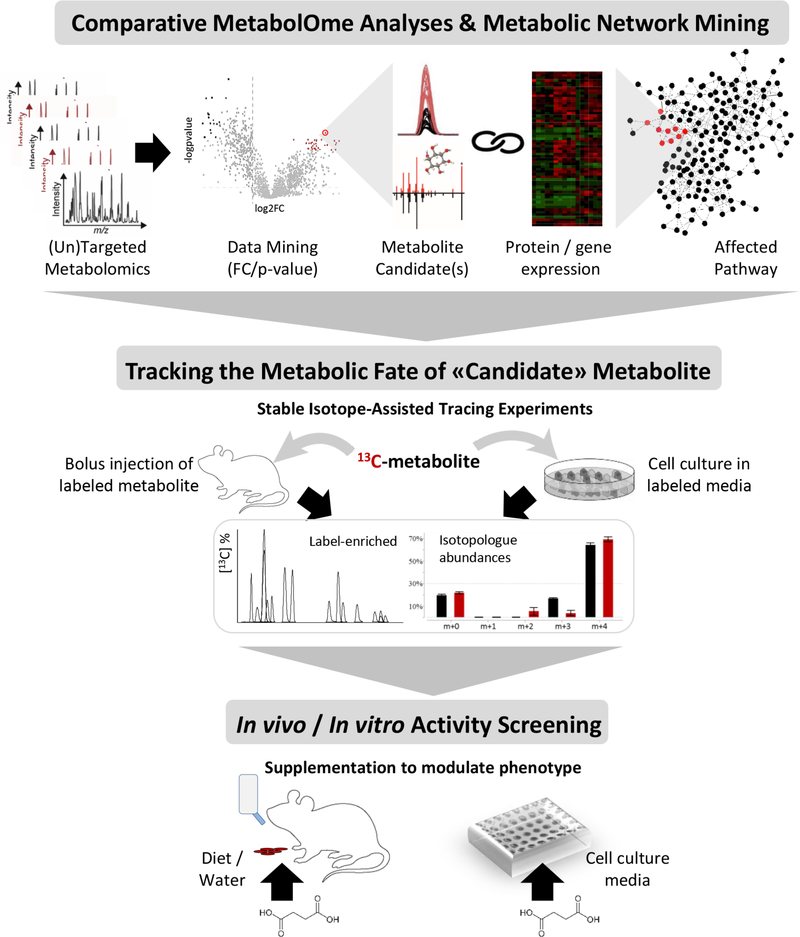
Figure 4.
Metabolomics-guided identification of bioactive metabolite candidate(s) followed by activity assessment. Workflow to elucidate the metabolite activity and role at the system’s level starts with the comparative metabolome analyses - a discovery-oriented untargeted or broad-scale targeted profiling analysis is followed by data mining to select the metabolite candidate(s) based on the significance (p-value), amplitude (fold change) and direction of its change (i.e. accumulation or depletion) in the studied system (e.g. cell media, mice/human plasma, etc.). To this end, statistical approaches are applied, and the metabolite is identified using spectral libraries (i.e. MS/MS matching). Protein and gene expression data (as a result of proteome and/or transcriptome analyses) are then used to support and help filter the candidate metabolites involved in the locally enriched part of the metabolic network, i.e. the same module or biochemical pathway. For example, the depletion of one a metabolite will usually be coupled by the significantly higher increased expression of its converting enzyme(s)) responsible for metabolite consumption. To gain further insights into the metabolic fate of the “metabolite-candidates” metabolite, the next step involves stable isotope-assisted (13C or 15N) tracing experiments to identify the active pathways used for metabolite catabolism. Different conditions can be tested in a longitudinal assay to gather the information about the metabolite uptake and conversions (i.e. label-enriched metabolites based on MS-based isotopologue abundance analyses, or NMR based flux analysis). Lastly, the metabolite supplementation experiments are performed using in vitro cell or organoid models, and in vivo models. These assays provide the direct information about how the phenotype is affected or modified via endogenous metabolite supplementation (ingestion through diet, drinking water or by addition to cell culture medium).