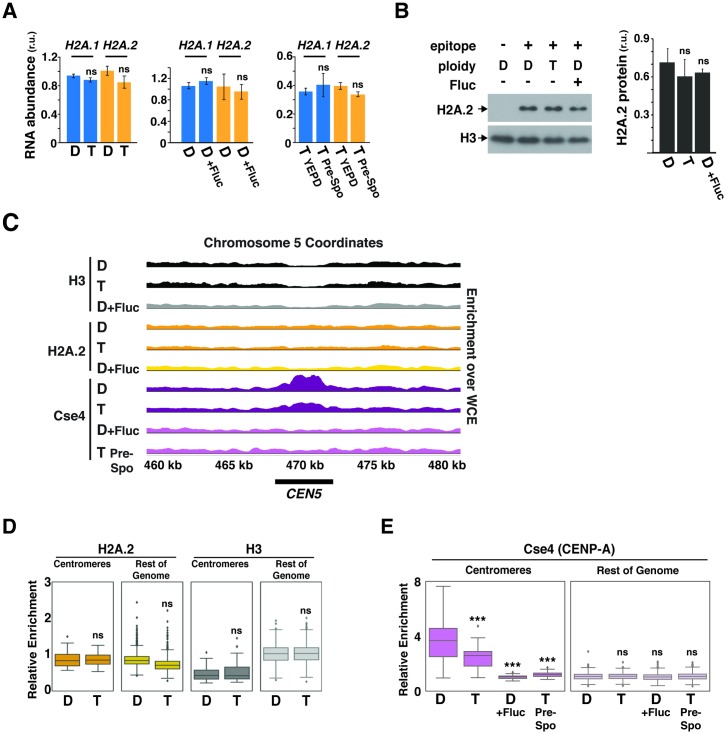
Fig 4. Impact of ploidy and environment on histone expression and deposition.
(A) RT-qPCR analysis of expression of H2A.1 and H2A.2 in the indicated genotypes and conditions. “D” represents a WT [11,22] diploid strain and “T” a WT [1111,2222] tetraploid strain. The average results for 3 biological replicates are presented after normalization to H3 mRNA, along with the standard deviation. There were no significant differences by one-way ANOVA. (B) Immunoblot of levels of H2A.2-FLAG in the indicated genotypes and conditions. Quantification using a LI-COR Odyssey imaging system is shown on the right. There were no significant differences by one-way ANOVA. (C) ChIP-seq analysis. Plots of bedgraphs of normalized read densities are shown for the indicated genotypes and conditions. Data represent the results of 1 replicate of each sample for CEN5. Data for all centromeres are shown in S6 Fig, and scatterplots of replicate data are shown in S7 Fig. (D and E) Boxplots showing normalized ChIP-seq enrichments for H2A.2 (D), H3 (D), and Cse4/CENP-A (E) (relative to the WCE sample; see Materials and methods) of nonoverlapping 5 kb tiles of the C. albicans genome separated into those overlapping with annotated centromeres versus the remainder of the genome. p-Values were determined using the Mann-Whitney U test. Cse4/CENP-A association with centromeres was significantly different in diploids versus tetraploids (p < 1 × 10−5), diploids propagated in YEPD versus YEPD plus fluconazole (p < 1 × 10−15), and tetraploids propagated in YEPD versus prespo (p < 1 × 10−13). Please see S1 Data for the numerical values summarized in (A). ChIP-seq data are available from the NCBI GEO website (accession number GSE122037). CENP-A, Centromere Protein A; ChIP-seq, chromatin immunoprecipitation with massively parallel DNA sequencing; Cse4, Centromeric Histone H3-like Protein; Fluc, fluconazole; GEO, Gene Expression Omnibus; H2A, Histone 2A; NCBI, National Center For Biotechnology Information; n.s., not significant; prespo, presporulation; RT-qPCR, reverse-transcription quantitative PCR; r.u., relative units; WCE, whole-cell extract; WT, wild type; YEPD, yeast extract peptone dextrose.