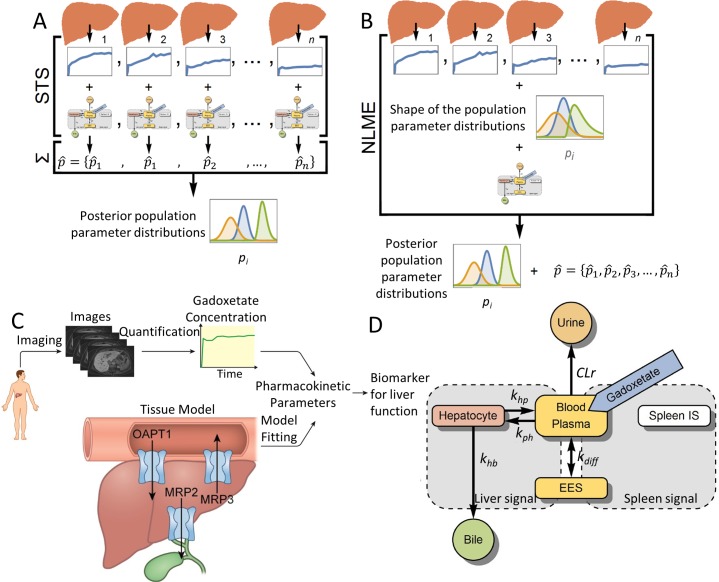
Fig 1. NLME, the mechanistic model framework, and model.
(A) The standard-two-stage (STS) method. The model is parametrized for each data set separately, and then the parameter values are combined to derive population parameter distributions. (B) The ‘non-linear mixed effect’ (NLME) method. The shapes of the population parameter distributions are first postulated, then distributions are parametrized to all datasets, and finally each patient is parametrized following the population parameter distributions with a joint-likelihood function. This allows NLME to use the global information obtained from an entire cohort, which is utilized to improve model parametrization for each individual subject. (C) The framework consists of gadoxetate-enhanced images, which are processed to obtain gadoxetate concentrations in the liver. A mechanistic systems pharmacology model, describing how gadoxetate is taken up and excreted, is fitted to the data using NLME parameterization to obtain reliable pharmacokinetic parameters, which can be used as biomarkers for liver function. (D) Schematic diagram of the mechanistic model for quantification of liver transporter function. Rounded rectangles represent compartments in the model, with arrows indicating gadoxetate fluxes between blood plasma and extracellular extravascular space (EES; kdiff), elimination via the kidneys to urine (CLr), uptake into hepatocytes (kph), back-flux from hepatocytes into blood plasma (khp), and excretion from hepatocytes into bile (khb). Gadoxetate injection into the blood-plasma compartment is indicated in blue. Gray areas show the signal part of the model in which compartmental gadoxetate concentrations are combined to predict the information in the gadoxetate-enhanced MRI time series.