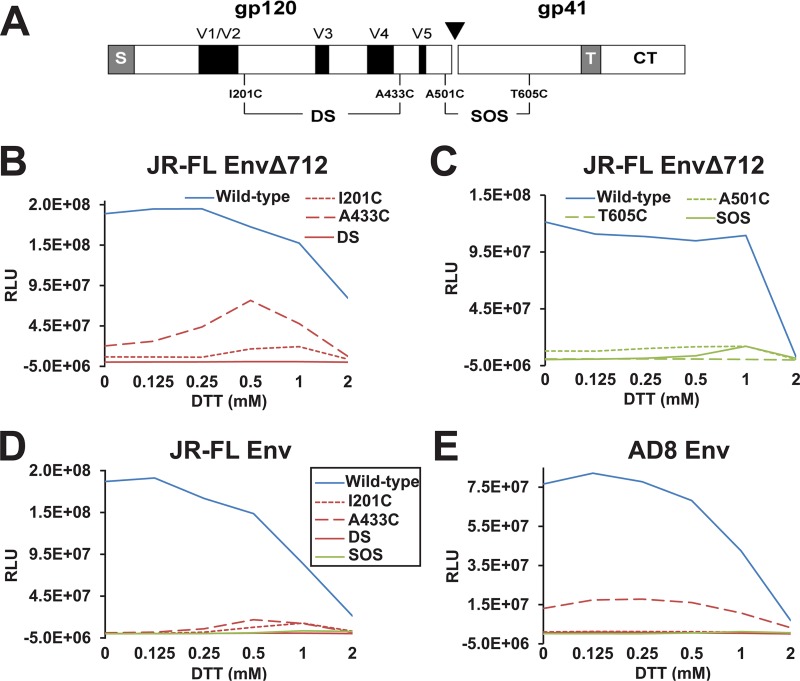
FIG 1.
Infectivity of recombinant viruses with HIV-1JR-FL and HIV-1AD8 Env variants. (A) Schematic of the HIV-1 Env, with the gp120-gp41 cleavage site depicted as a black triangle. The gp120 major variable regions (V1 to V5) are shown as black boxes. The cysteine substitutions associated with the DS and SOS mutants are indicated. S, signal peptide; T, transmembrane region; CT, cytoplasmic tail. (B to E) Recombinant luciferase-expressing viruses with the indicated Envs were incubated with Cf2Th-CD4/CCR5 cells in the presence of 0 to 2 mM dithiothreitol (DTT). The levels of the p24 Gag protein in the virus preparations were similar for the different Env variants (Fig. 2A). Forty-eight hours later, the Cf2Th-CD4/CCR5 target cells were lysed, and the luciferase activity was measured. The reported relative light unit (RLU) values reflect the infectivities observed for each Env mutant. The y axis has been scaled so that the RLU values near the background of the assay can be visualized. The data are representative of those obtained in at least three independent experiments. The key in panel D also applies to panel E.