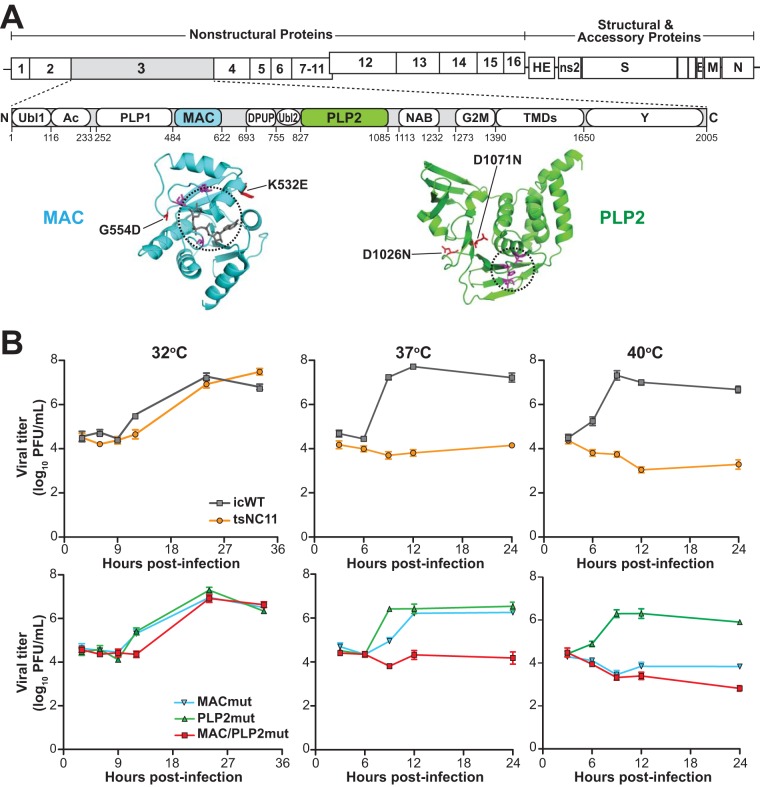
FIG 1.
Evaluation of the replication kinetics of coronavirus temperature-sensitive mutants at permissive and nonpermissive temperatures. (A) Schematic diagram of the MHV genome and the domains of nsp3. Abbreviations: Ubl1, ubiquitin-like domain 1; Ac, acidic region; PLP1, papain-like protease 1; MAC, macrodomain; DPUP, domain preceding Ubl2 and PLP2; Ubl2, ubiquitin-like domain 2; PLP2, papain-like protease 2; NAB, nucleic acid-binding domain; G2M, coronavirus group 2 marker domain; TMDs, transmembrane domains; Y, coronavirus highly conserved domain. Representative structures of the macrodomain with ribose (229E; PDB 3EWR) and PLP2 (MHV; PDB 4YPT) are shown in cyan and green with catalytic pockets circled, and the residues involved in catalysis are shown in magenta. The mutations described in this study are shown in red. (B) Growth kinetics of MHV and mutants at three temperatures. DBT cells were inoculated with the indicated virus (at a multiplicity of infection [MOI] of 5) for 1 h at 37°C and then shifted to the indicated temperatures. Culture supernatants were collected at the indicated hours postinfection and titrated in DBT cells at 32°C. The data are representative of two independent experiments. Error bars indicate ± the standard deviation (SD).