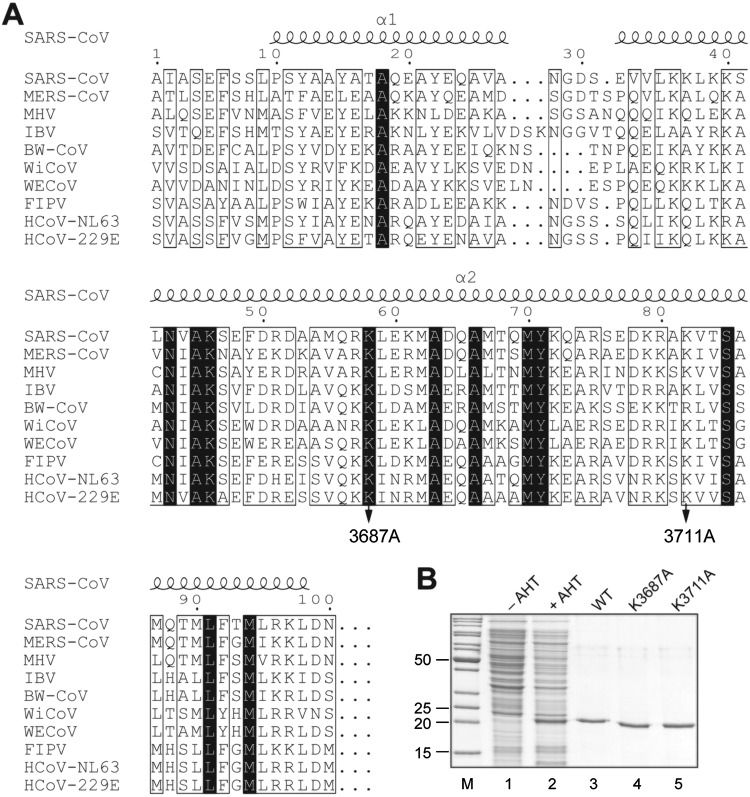
FIG 1.
Production of HCoV-229E nsp8 in E. coli. (A) Multiple-sequence alignment of the N-terminal regions of coronavirus nsp8 proteins representing the genera Alpha-, Beta-, Gamma-, and Deltacoronavirus. Sequences were aligned using the Clustal Omega program (62) and converted using the ESPript program (63). The black background indicates invariant residues. The alanine substitutions in HCoV-229E nsp8 generated in this study are indicated by arrows. Numbers indicate the positions in the HCoV-229E polyprotein 1a/1ab sequence. SARS-CoV, severe acute respiratory syndrome coronavirus (GenBank accession number AY291315); MERS-CoV, Middle East respiratory syndrome coronavirus (GenBank accession number JX869059.2); MHV, mouse hepatitis virus A59 (GenBank accession number NC_001846.1); IBV, avian infectious bronchitis virus (GenBank accession number NC_001451.1); BW-CoV, beluga whale coronavirus SW1 (GenBank accession number NC_010646); WiCoV, wigeon coronavirus HKU20 (GenBank accession number JQ065048); WECoV, white-eye coronavirus HKU16 (GenBank accession number JQ065044); FIPV, feline infectious peritonitis virus (GenBank accession number NC_002306); HCoV-NL63, human coronavirus NL63 (GenBank accession number NC_005831); HCoV-229E, human coronavirus 229E (GenBank accession number NC_002645). Secondary structure elements determined by crystal structure analysis of SARS-CoV nsp8 (chain H; PDB accession number 2AHM) (13) are shown above the alignment. (B) Coomassie brilliant blue-stained 12% SDS-polyacrylamide gel showing the production and purification of HCoV-229E nsp8-CHis6. Lanes 1 and 2, total lysates of E. coli TB1(pCG1) cells transformed with pASK-Ub-nsp8-CHis6 plasmid DNA and grown in the absence (–) or presence (+) of anhydrotetracycline (AHT); lanes 3 to 5, purified nsp8-CHis6 proteins (wild type [WT] or variants containing the indicated alanine substitutions of conserved residues). Lane M, marker proteins (molecular masses [in kilodaltons] are indicated to the left).