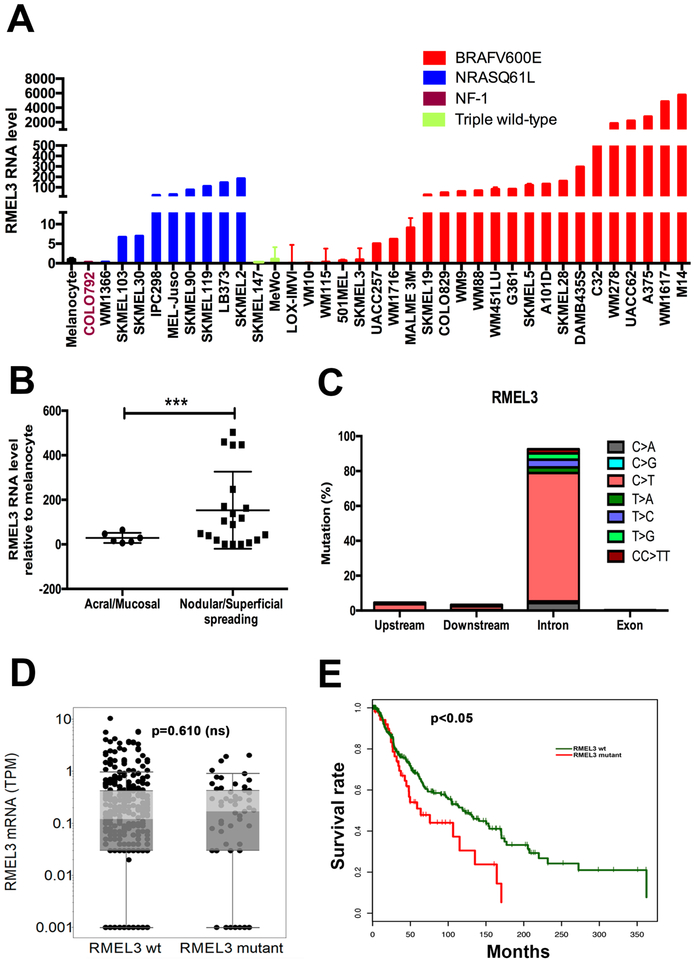
Figure 1. High RMEL3 expression predominates in BRAFV600E followed by NRAS mutant melanoma cell lines, as well as in nodular/ superficial spreading in comparison with acral/mucosal melanomas.
(A) RT-qPCR analysis of RMEL3 RNA levels in melanocytes and melanoma cell lines of indicated genotype regarding major câncer drivers (BRAF, NRAS, and NF1). (B) RT-qPCR analysis of RMEL3 expression in melanoma subtypes: acral/mucosal and nodular/superficial spreading cutaneous melanoma, for which patient specimens were obtained from the Barretos Cancer Hospital, São Paulo state, Brazil. In A and B, relative expression was calculated according to 2−ΔΔCT method usingTBP (Tata-box binding protein) as endogenous control, and the mean levels of RMEL3 RNA in different foreskin human melanocyte primary cultures were used as reference in both A and B, totaling the use of four different melanocyte cultures derived from different donors. Error bars represent SEM of 3 independent experiments. ***p < 0.005. Asterisks indicate statistically significant differences between groups based on Fisher’s exact test. (C) Frequency of base substitution mutations in the RMEL3 locus from a total of 579 base substitutions reported by ICGC in a set of 129 melanoma samples. (D) RMEL3 expression levels (TPM) grouped according to the occurrence (RMEL3 mutant) or not (RMEL3 wt) of somatic mutation in the RMEL3 gene detected in the TCGA melanoma dataset of 450 samples. (E) Survival curves for melanoma patients of the TCGA database carrying (RMEL3 mutant) or not (RMEL3 wt) mutations in the RMEL3 gene. Statistical analyses used chi-squared test in D and E