Figure 3.
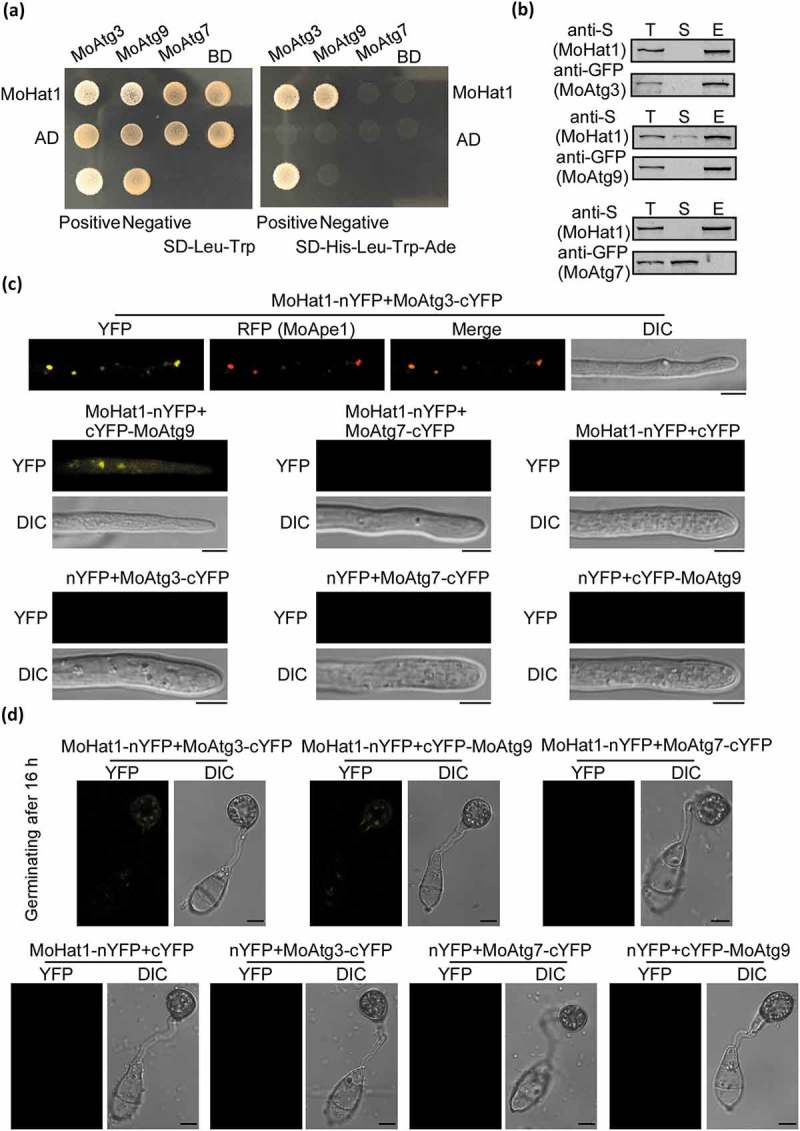
MoHat1 interacts with MoAtg3 and MoAtg9. (a) Yeast two-hybrid analysis for the interaction between MoHat1 and Atg proteins. The pGADT7 and pGBKT7 fused with specific genes were co-introduced into the yeast AH109 strain, and transformants were plated on SD-Leu-Trp as control and on selective SD-Leu-Trp-His-Ade for 5 days. The pair of plasmids pGBKT7-Lam and pGADT7-T, empty pGADT7 and pGBKT7 were used as the negative controls. MoAtg7 was used also as a negative control that had no interaction with MoHat1. (b) Co-IP assays. Western blots of total proteins, suspensions and proteins eluted from anti-S agarose from transformants co-transformed MoAtg3-GFP and MoHat1-S, MoAtg9-GFP and MoHat1-S, and also MoAtg7-GFP and MoHat1-S were detected with anti-S or anti-GFP antibodies. T, total; S, suspensions; E, elution. (c and d) BiFC assay for the patterns of MoHat1-MoAtg3, MoHat1-MoAtg9 and MoHat1-MoAtg7 in vivo. Both aerial hypha tips and 16 h appressoria were examined by DIC and fluorescence microscopy. Strains expressing MoHat1-nYFP and MoAtg7-cYFP, MoHat1-nYFP and empty cYFP, MoAtg3-cYFP and empty nYFP, MoAtg9-cYFP and empty nYFP, MoAtg7-cYFP and empty nYFP were used as negative controls. MoApe1-RFP was used as a phagophore assembly site (PAS) marker. Scale bar: 10 μm.
