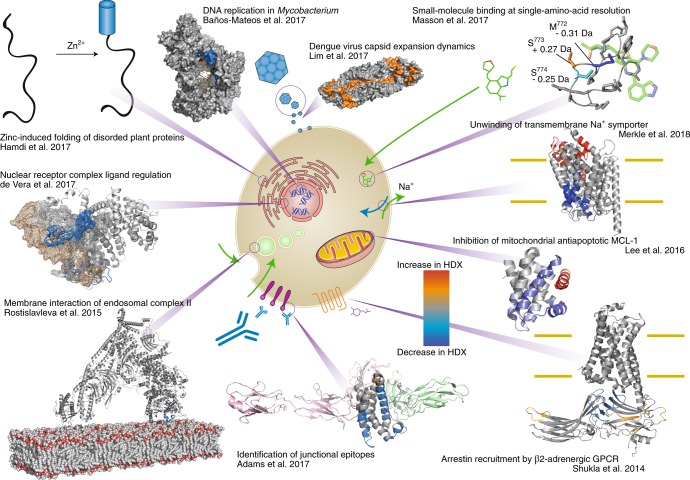
Fig. 1. The wide range of applications for HDX-MS in many protein-folding studies.
Clockwise from top left: Hamdi et al.24 localized dehydration and zinc-activated disorder-to-order transitions in abiotic plant stress response proteins using HDX-MS. Baños-Mateos et al.25 demonstrated how HDX-MS can be used in combination with X-ray crystallography and cryo-electron microscopy (cryo-EM) when determining the mechanism of exonuclease activity of DnaE1. Lim et al.26 determined how an increase in temperature alters the Dengue virus capsid structure, resulting in an alteration in antibody-binding mode. Using ETD-HDX/MS/MS, Masson et al.27 discerned the basis of isotype specificity of pharmaceutical compounds by determining single-residue exchange rates. Merkle et al.28 localized the substrate-dependent partial unwinding of transmembrane helices, which facilities substrate translocation using HDX-MS. Lee et al.29 revealed a new allosteric mechanism for interrupting the antiapoptotic binding of MCL-1 to BH3 domains, providing a new avenue for cancer therapy. Shukla et al.30 demonstrated how HDX-MS can provide mechanistic and dynamic detail to cryo-EM structures, and how HDX-MS can aid modeling of X-ray structures within the cryo-EM density. Adams et al.31 illustrated the utility of HDX-MS, used in conjunction with X-ray crystallography and biophysical methods, to reveal how the monoclonal antibody VHH6 contemporaneously interacts with IL-6 and gp80 through a junctional epitope. Rostislavleva et al.32 pushed the limits of HDX-MS with the large lipid kinase VPS34 complex II by both determining the membrane-interacting regions of the lipid kinase and screening nanobodies to facilitate crystallization and subsequent structure determination. By altering the pH of the labelling solution, de Vera et al.33 observed interactions of disordered protein domain on a millisecond timescale by HDX-MS.