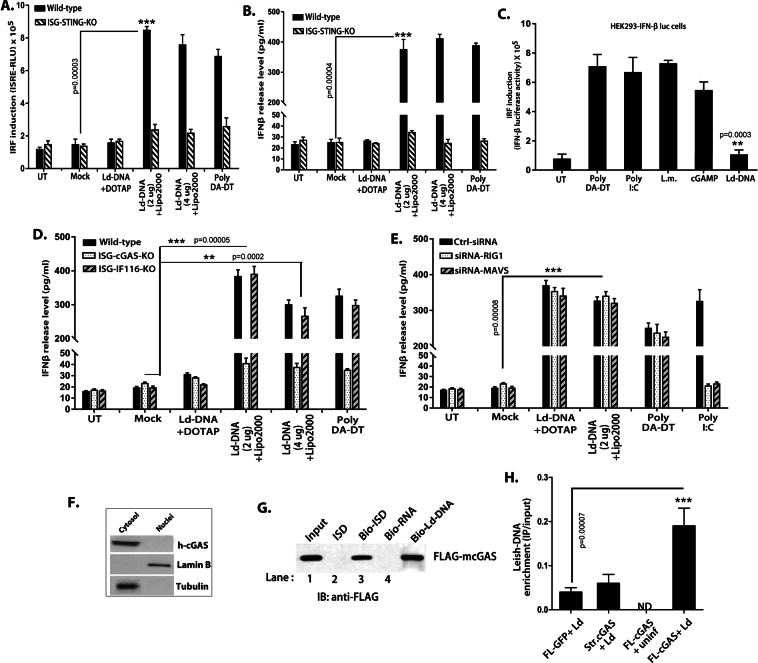
Figure 5.
Cytosolic DNA sensor cGAS is targeted by Ld-DNA for for IFN-β activation. (A,B) Mouse RAW264.7 reporter derived STING ablated [STING KO] cells and control cells [ISG-WT] were mock transfected or with Ld-DNA [2.5 ug/ml with DOTAP or Lipofectamine 2000]. IRF pathway activation was measured by luciferase reporter assay (A) and production of IFN-β was estimated at the release level in the culture supernatant by ELISA (B). (C) HEK293- IFN-β-luciferase reporter cells were transfected with poly(DA-DT) or poly(I:C) or Ld-DNA or treated with cGAMP or infected with L. monocytogenes. After overnight stimulations, IRF induction was measured through luciferase activity. (D,E) Mouse RAW264.7 reporter derived cGAS ablated [cGAS KO] cells or IFI16 ablated [IFI16 KO] and control cells [ISG-WT] (D) or pre-transfected with with targeted RIG-1-siRNA or MAVS-siRNA or non-targeted (NT) control siRNA (E) were employed. These cells were mock transfected or with Ld-DNA [2.5 ug/ml with DOTAP or Lipofectamine 2000] or poly (DA-DT). Production of IFN-β was estimated at the release level in the culture supernatant by ELISA (D,E). (F) THP-1 cells were transfected with Ld-DNA [2.5 ug/ml with Lipofectamine 2000]. The cytosolic and nuclear fractions were prepared after 8 hrs post transfection. The fractions were immunoblotted with the indicated antibodies. (G) FLAG-m-cGAS was expressed and purified from E.coli. Subsequently, the FLAG-tagged cGAS was incubated with ISD or biotinylated-ISD or biotinylated-RNA in the presence streptavidin beads [See method section]. The bound proteins were then eluted in SDS sample buffer for immunoblotting with an anti-FLAG antibody. (H) HEK293 cells stably expressing FLAG-tagged constructs of mouse cGAS [HEK293-FLAG-m-cGAS and HEK293-FLAG-m-strep-cGAS] were infected with L.donovani parasites (parasite: macrophage = 10: 1) and crosslinked with 4% PFA. The m-cGAS was then precipitated with anti-FLAG antibody. The DNA in the precipitate was then subjected to qRT-PCR of L.donovani specific kDNA sequence. Quartiles were normalized to the inputs. Data represent mean ± SEM of at least three independent experiments (n = 4 for A–C and G; and n = 3 for D–F and H). Statistically significant results are marked as **p < 0.001 and ***p < 0.0001.