Abstract
Background: Accumulating studies have demonstrated that some long non-coding RNAs (lncRNAs) play critical roles in the pathogenesis of atherosclerosis. We aimed to identify circulation lncRNAs that are potential biomarkers to evaluate coronary atherosclerotic plaque stability. Methods and Results: The transcriptomes of blood samples of three patients with stable plaque and three patients with unstable plaque were sequenced by RNA-sequencing. A total of 62 lncRNAs were found to be differentially expressed in patients with unstable plaques. The expressions of four candidate lncRNAs (ANP32A-005, TULP4-005, PDCD4-010, and SNHG7-003) were quantified using blood samples from 15 patients with stable plaques and 15 patients with unstable plaques, subsequently. In addition, the expression levels of these four lncRNAs in LPS (lipopolysaccharide)-activated THP-1 monocytes and THP-1-derived macrophages were measured. LncRNA-SNHG7-003 was validated to be significantly down-regulated in blood samples of patients with unstable plaques and LPS-stimulated monocytes and macrophages. Moreover, plasmid-transfection mediated over-expression of SNHG7-003 markedly inhibited the activation of NF-κB pathway, and reduced the secretion of inflammatory mediators (TNF-α, IL-1β, MCP-1 and MMP-9) in LPS-activated THP-1 monocytes and macrophages. Conclusion: LncRNA-SNHG7-003 inhibits NF-κB activation and regulates inflammatory responses in human monocytes and macrophages. Blood lncRNA-SNHG7-003 is a potential biomarker for evaluating plaque stability in patients with coronary artery diseases.
Keywords: Atherosclerosis, plaque stability, long non-coding RNA, SNHG7-003
Introduction
Coronary artery unstable plaque rupture and thrombosis are the most important pathological basis of acute coronary syndrome (ACS), which is life threatening to patients with coronary artery diseases (CAD) [1]. Even though sometimes unstable plaques might not cause severe arteriostenosis, they could lead to serious cardiac events after rupture. Many patients with coronary artery diseases do not demonstrate evident clinical symptoms, but they have high risk of acute coronary events [2,3]. Rupture of unstable plaques can lead to myocardial infarction and is the leading cause of morbidity and mortality. Postmortem pathological examinations showed that 60% to 70% of the culprit lesions for ACS demonstrated plaque rupture [4-6], and several studies have revealed the presence of plaque rupture in 14% to 66% of the culprit lesions from patients with ACS in vivo [7-9]. It has been reported that only a few patients, who have suffered from acute myocardial infarction (AMI), are clinically categorized into high risk population before AMI [10]. Hence, early recognition of unstable plaques would be of great value for patients with CAD.
Clinically, Intravascular Ultrasound (IVUS) and its derivatives, as well as Optical coherence tomography (OCT) could provide clinicians with morphological features of coronary artery atherosclerotic plaques, which could help with plaque stability evaluation [11-13]. However, these tools are too invasive and expensive to be widely used for screening of unstable plaques. Strategies targeting circulation biomarkers, such as long non-coding RNAs (lncRNAs), would provide easier ways to evaluate plaque stability in patients with CAD. Identified as non-protein-coding RNAs with longer than 200 nucleotides in 1992 [14,15], lncRNAs were found to regulate many biological processes under both physiological and pathological conditions [16,17]. They are able to regulate gene expression and functions of other molecules through multiple approaches, such as epigenetic regulation, transcriptional regulation, post-transcriptional regulation, molecular sponges, molecular chaperones [18-21]. Some cardiovascular lncRNAs including Novlnc6, Mhrt, MALAT1 and Tie-1-AS, have also been identified to play a role in cardiovascular diseases like AMI and myocardial hypertrophy [22].
Recently, lncRNAs have been demonstrated to be potential key regulators of the inflammatory responses [23]. Since the pivotal roles of inflammation in the pathogenesis of atherosclerosis have been recognized and proved [24-26], this implies a potential link between lncRNA expression and incidence of atherosclerosis. Indeed, some studies have already revealed this correlation. One of the most studied lncRNAs, ANRIL, is an antisense non-coding RNA located in the INK4 locus, which is adjacent to chromosome 9p21 locus (Chr9p21) [22], a strong genetic risk factor for CAD [27,28]. Further studies demonstrated that lncRNA-ANRIL expression was observed in atherosclerotic tissues and atherogenic cells, such as endothelial cells, smooth muscle cells, and monocyte-derived macrophages [29], and that the expression level of ANRIL was correlated with the severity of atherosclerosis [27,30]. In addition, MIAT, a lncRNA identified by Ishil et al., was reported to be associated with myocardial infarction and regulate the functions of endothelial cells [31,32]. Expression levels of MIAT were reported to be reduced in blood mononuclear cells of patients with ST segment-elevation myocardial infarction [33]. Moreover, LncRNA-p21 has been considered to play a role in atherogenesis as it can regulate the proliferation and apoptosis of smooth muscle cells, as well as neointimal hyperplasia [34]. However, whether lncRNAs can be used as reliable circulation biomarkers to distinguish stable and unstable plaque in patients with CAD has not been systematically explored so far.
In this study, we aimed to identify circulation lncRNA molecules related to the coronary atherosclerotic plaque stability by RNA-sequencing (RNA-Seq) of blood samples from patients with CAD, and verified some of the top differentially expressed lncRNAs by RT-PCR. A gain-of-function study was performed to explore the role of validated lncRNA in atherosclerosis-related inflammation in a human monocytic cell line THP-1 and THP-1 derived macrophages through plasmid-mediated over-expression of target gene.
Materials and methods
Study design and ethic statement
Peripheral blood was collected from sex and age matched patients with CAD, in which 3 patients were diagnosed with stable coronary atherosclerotic plaque and 3 patients with unstable plaque. Transcriptomes of these blood samples were sequenced by RNA-Seq to screen out candidate lncRNA molecules that might be related to plaque stability. In order to validate the preliminary results, the expressions of four candidate lncRNAs were quantified in a lager sample size containing blood samples from 15 patients with stable plaques and 15 patients with unstable plaques. In addition, the expression levels of these 4 candidate lncRNAs were quantitated in LPS (lipopolysaccharides)-induced THP-1 monocytes and THP-1-derived macrophages. Over-expression of validated lncRNA in THP-1 monocytes and THP-1-derived macrophage was conducted to explore its role in atherosclerosis-related inflammation. This study was approved by the ethics committee of Shanghai Chest Hospital, China. The study protocol was carefully explained to the participants and participation was fully voluntary. Written informed consent was obtained from all participants.
Patient selection
Patients meeting the following criteria were assigned to the unstable plaque group: (1) Clinically diagnosed as acute coronary syndrome (namely unstable angina, acute ST-segment elevation myocardial infarction, or acute non-ST-segment elevation myocardial infarction); (2) Obvious atherosclerotic lesions with at least 50% diameter stenosis or thrombosis were showed when performing coronary angiography; (3) Intravascular ultrasound indicated that the lesions were unstable coronary atherosclerotic plaques (namely plaque with ulceration, thrombosis, or broken fibrous cap; lipid core > 1 mm2 or lipid core to plaque ratio > 20%; fibrous cap < 0.7 mm, as previously defined [35]).
Patients meeting the following criteria were assigned to the stable plaque group: (1) Clinically diagnosed as stable angina pectoris; (2) One and only one atherosclerotic lesion with at least 50% diameter stenosis showed when performing coronary angiography; (3) Intravascular ultrasound indicated that the lesion was a stable coronary atherosclerotic plaque (namely plaque without ulceration, thrombosis, and broken fibrous cap; lipid core ≤ 1 mm2 or lipid core to plaque ratio ≤ 20%; fibrous cap ≥ 0.7 mm).
Patients with chronic active inflammatory disease, malignant tumor, blood disease, autoimmune disease, and severe hepatic or renal dysfunction were not included. The clinical characteristics of patients whose peripheral blood samples were used for lncRNAs validation are listed in Table 1.
Table 1.
The clinical characteristics of patients whose peripheral blood samples were used for lncRNAs validation
| Stable plaque Group (n = 15) | Unstable plaque Group (n = 15) | P value | |
|---|---|---|---|
| Age | 64.27 ± 5.32 | 62.00 ± 10.99 | 0.481 |
| Male | 13 (86.7%) | 14 (93.3%) | 0.543 |
| Hypertension | 9 (60%) | 9 (60%) | 1.000 |
| Diabetes mellitus | 2 (13.3%) | 4 (26.7%) | 0.361 |
| Smoke | 3 (20%) | 3 (20%) | 1.000 |
| Dyslipidemia | 11 (73.3%) | 6 (40.0%) | 0.065 |
| TG | 2.35 ± 1.50 | 1.31 ± 0.93 | 0.039 |
| TC | 4.00 ± 1.17 | 4.32 ± 1.15 | 0.481 |
| HDL | 1.04 ± 0.26 | 0.99 ± 0.33 | 0.661 |
| LDL | 2.44 ± 0.78 | 2.81 ± 0.82 | 0.235 |
| cTNI | 0.01 ± 0.01 | 27.76 ± 36.30 | 0.010 |
| LVEF | 63.86 ± 2.71 | 56.50 ± 12.08 | 0.043 |
| Gensini | 7.16 ± 3.81 | 65.21 ± 37.96 | < 0.001 |
TG, Plasma Total Triglyceride; TC, Plasma Total Cholesterol; HDL, Plasma High Density Lipoprotein; LDL, Plasma Low Density Lipoprotein; cTNI, Plasma Cardiac Troponin I; LVEF, left ventricular ejection fraction.
Blood collection and total RNA isolation
Blood samples were collected with EDTA-anticoagulant tubes from patients with CAD right after their diagnosis. After collection, 1 ml fresh blood was added immediately to a clean tube containing 2 ml RNAzol® BD (Molecular Research Center, Inc, USA) and 27 μl acetic acid (Macklin, Inc, China), with vigorously shaking for 30 seconds. A total of 36 samples, comprising 18 samples in the stable plaque group and 18 samples in the unstable plaque group, were collected and stored at -80°C until use. Blood total RNA was extracted using RNAzol® BD (Molecular Research Center, Inc, USA) by following the manufacturer’s instructions and was dissolved in RNase-free water. The purity and concentration of the extracted total RNA was tested with NanoDrop ND-1000 (Thermo Scientific, USA). RNA integrity was validated with 1% agarose gel electrophoresis. Three RNA samples in each group from age and sex matched patients were selected for transcriptome sequencing analysis, and the remaining 30 samples were used for subsequent PCR verification.
Preparation of cDNA libraries and sequencing
1~2 μg total RNA of each sample was used to prepare cDNA libraries, as previously described [36]. Briefly, the RiboZero Magnetic Gold Kit (Human/Mouse/Rat) (Illumina, Inc, USA) was used to remove ribosomal RNA from each sample. Then libraries preparation, including steps like RNA fragment, radom primed 1st strand cDNA synthesis, dUTP based 2nd strand cDNA synthesis, adaptor ligation, and PCR amplification, was conducted using KAPA Stranded RNA-Seq Library Prep Kit (Illumina, Inc, USA). The quality of prepared libraries was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Finally, TruSeq SR Cluster Kit v3-cBot-HS (Illumina, Inc, USA) and Illumina HiSeq 4000 instrument were used to conduct sequencing. All procedures were strictly in accordance with manufacturers’ instructions.
Analysis of RNA-Seq data
Raw sequencing data were quality controlled and pre-processed, and were aligned with human reference transcriptome (version GRCh37.p13) using Hisat2 (version 2.0.2). Total transcripts were assembled using StringTie (version 1.3.1c). Then the computation of transcripts abundance and screening of differentially expressed transcripts were conducted with the help of Ballgown (version 2.8.4). Transcripts with fold change ≥ 1.5 and P value < 0.05 between the stable plaque group and the unstable plaque group were considered as differentially expressed transcripts. Hierarchical Clustering was conducted with R (version 3.4.1) to clarify the expression patterns of distinguishable transcripts.
Cell culture and treatment
THP-1 cells (purchased from the Cell Bank of Chinese Academy of Sciences, China) were cultured in RPMI 1640 medium (containing HEPES; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Biological Industries, USA) and 0.05 mM β-mercaptoethanol (Sigma-Aldrich, USA), in an atmosphere of 95% air and 5% CO2 at 37°C. THP-1 monocytes were stimulated with 160 nM PMA (phorbol-12-myristate acetate; Sigma-Aldrich, USA) for 24 h, in order to induce the differentiation of THP-1 monocytes into macrophages.
Plasmid vector (pcDNA3.1+) with enzymatically ligated full length sequence of SNHG7-003 was ordered from Sangon Biotech Co., Ltd., Shanghai, China. Lipofectamine® 2000 Reagent (Invitrogen, USA) was used for transfection of pcDNA3.1+/SNHG7-003 recombinant plasmid or pcDNA3.1+ control plasmid into THP-1 monocytes and macrophages. At 24 hours (THP-1 monocytes) and 30 hours (THP-1 derived macrophages) after plasmid transfection, cells were stimulated with 1 μg/ml LPS (Beyotime, China). After LPS stimulation for 2 hours, cells were harvested for western blot. After LPS stimulation for 24 hours, culture supernatant was harvested for cytokine assay, while cells were harvested for RNA isolation and PCR.
Reverse transcription and quantitative PCR (RT-qPCR)
Real-time fluorescence quantitative polymerase chain reaction assays were used to detect relative expression levels of candidate lncRNAs in blood samples and in LPS-stimulated THP-1 monocytes and THP-1-derived macrophages. Blood total RNA was isolated as mentioned above. Total RNA from THP-1 cells was isolated using RNAiso Plus (Takara Biomedical Technology Co., Ltd, Beijing, China). DNase I (Beyotime, China) was used to remove DNA from each RNA sample. After that, cDNA was synthesized from each purified RNA sample using the M-MuLV First Strand cDNA Synthesis Kit (Sangon Biotech, China). Real-time quantitative PCR was performed using 2×SYBR Green qPCR Master Mix (Bimake, USA), with the Applied Biosystems® 7500 Real-Time PCR Systems. All procedures were strictly in accordance with manufacturers’ instructions. Expression levels of candidate lncRNAs were calculated by the ΔΔCT method [37], with normalization to GAPDH. The sequences of primers are shown in Table 2.
Table 2.
The sequences of DNA primers used for RT-qPCR in this study
| Transcripts | Forward | Reverse |
|---|---|---|
| GAPDH | 5’-TGGACCTGACCTGCCGTCTA-3’ | 5’-GGAGTGGGTGTCGCTGTTGA-3’ |
| ANP32A-005 | 5’-ATCGCTCTGGTGTTTGCTCTT-3’ | 5’-GGCTGCTGTCCTTGGCTTC-3’ |
| TULP4-005 | 5’-CTCCCAGCCGTGAATAATCT-3’ | 5’-ACAAGAATGAGCCTGTCTTCC-3’ |
| PDCD4-010 | 5’-ATCACCTTGTTAAAGAGGCTATTA-3’ | 5’-CGCTCCAGCACAGAGTATGA-3’ |
| SNHG7-003 | 5’-CTTCGCCTGTGATGGACTTC-3’ | 5’-CCTGCCCATCCCTTTATTCC-3’ |
Western blot
NF-κB activation was evaluated by measuring the phosphorylation of NF-κB p65 (S536) through western blot analysis. Cells were lysed in the RIPA (Radio-Immunoprecipitation Assay) lysis buffer (Beyotime, China), supplemented with the Protease Inhibitor Cocktail (Sigma-Aldrich, USA) and Phosphatase Inhibitor Cocktail (Sigma-Aldrich, USA), on ice for 15 minutes. After centrifugation at 12,000 g for 15 minutes at 4°C, whole-cell proteins in the supernatants were collected and quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, USA). Each protein sample was mixed with 5×SDS-PAGE Protein Loading Buffer (Beyotime, China), and boiled for 5 minutes. Equal amount (12 μg) of each protein sample was separated using 10% SDS-PAGE and transferred to PVDF membrane (Millipore, USA). The membranes were blocked for 30 minutes in QuickBlock™ Blocking Buffer for Western Blot (Beyotime, China), and then incubated with primary antibodies at 4°C overnight. The primary antibodies were as follows: rabbit anti-NF-κB p65 antibody (Abcam, UK), rabbit anti-phosphorylated NF-κB p65 (Ser536) antibody (Abcam, UK), rabbit anti-GAPDH polyclonal antibody (Sangon Biotech, China). After incubation overnight, the membranes were washed with TBST Buffer (Sangon Biotech, China) for 3 times, and then incubated with HRP-linked anti-rabbit IgG antibody (Cell Signaling Technology, USA) at room temperature for 2 hours. Afterwards, the membranes were washed again with TBST for 3 times, and then developed using Tanon™ High-sig ECL Western Blotting Substrate (Tanon, China) and Tanon 4600 Automatic Chemiluminescence Imaging System (Tanon, China). The band intensities were quantified by densitometry using ImageJ software (version 1.0, National Institutes of Health, Bethesda, MD, USA), and were normalized to the expression levels of GAPDH.
ELISA (enzyme-linked immunosorbent assay)
The inflammatory responses of THP-1 monocytes and THP-1-derived macrophages were estimated by measuring the levels of inflammatory mediators, including TNF-α (tumor necrosis factor-α), IL-1β (interleukin-1β), MCP-1 (monocyte chemotactic protein 1), and MMP-9 (matrix metallopeptidase 9), secreted in cell culture supernatants through ELISA. ELISA kits used in this study were as follows: Human TNF-α ValukineTM ELISA Kit (Novus Biologicals, USA), Human IL-1β/IL-1F2 ValukineTM ELISA Kit (Novus Biologicals, USA), Human MMP-9 ValukineTM ELISA Kit (Novus Biologicals, USA), Human MCP-1ELISA Kit (Elabscience Biotechnology Co., Ltd, China). All experiments were performed according to the manufacturers’ instructions.
Statistics
Statistical analyses were performed using the SPSS software (version 23.0; IBM, USA). Results were presented as mean ± SD (standard deviation), and comparisons between groups were performed using paired t-test or unpaired t-test where appropriate. P value < 0.05 was considered statistically significant.
Results
Screening of differentially expressed lncRNAs between the stable plaque group and the unstable plaque group by RNA-Seq
In order to screen for potential lncRNAs that are associated with the plaque stability of coronary atherosclerosis, we performed RNA-Seq analyses of blood samples from patients. Totally, 28325 transcripts were identified in blood samples of patients in the stable plaque group, while 28489 transcripts were identified in the unstable plaque group. As shown in Figure 1, 908 transcripts were found differentially expressed between the two groups. Among them, 462 transcripts, including 119 lncRNAs and 343 protein-coding RNAs, were up-regulated in the unstable plaque group, while 446 transcripts, including 112 lncRNAs and 334 protein-coding RNAs, were down-regulated in the unstable plaque group. Hierarchical clustering of these differentially expressed transcripts showed that patients with unstable plaques could be clearly distinguished from patients with stable plaques based on centered Pearson correlation (Figure 2). A total of 62 differentially expressed lncRNAs (fold change ≥ 2) between the stable plaque group and the unstable plaque group are listed in Table 3. We therefore designed a series of primers for the top 5 most up-regulated and top 5 most down-regulated lncRNAs and attempted to verify their expression by qPCR. However, only the primers for ANP32A-005, TULP4-005, PDCD4-010, and SNHG7-003 were proven to be specific and effective. Hence, these 4 lncRNAs with valid qPCR primers were selected as candidate lncRNAs for subsequent investigations.
Figure 1.

Volcano plot showing the differences of transcripts expression between blood samples of patients with stable plaques and those with unstable plaques. Fold change (unstable/stable) and statistical significance (p value) were two parameters used to screen out differentially expressed transcripts in the volcano plot. X-axis represents log2 (Fold Change); Y-axis represents -log10 (p value). Two vertical lines represent threshold of up-regulated transcripts (right) and down-regulated transcripts (left), respectively (Fold change ≥ 1.5). The horizontal line represents threshold of statistical significance (p value ≤ 0.05). Red spots represent 462 up-regulated transcripts in the unstable plaque group; green spots represent 446 down-regulated transcripts in the unstable plaque group; grey spots represent 32511 transcripts without differential expression between the two groups.
Figure 2.

Heat map showing hierarchical clustering of differently expressed transcripts between blood samples of patients with stable plaques and those with unstable plaques. Each column represents a sample; each row represents a transcript. The left three samples are in the unstable plaque group; the right three samples are in the stable plaque group. Transcripts expressions are depicted using a color scale. Red indicates up-regulated, while green indicates down-regulated.
Table 3.
List of differentially expressed lncRNAs in blood samples of patients from the unstable plaque group and the stable plaque group
| Up regulated | Down regulated | ||||
|---|---|---|---|---|---|
|
|
|
||||
| lncRNA | Fold change | P value | lncRNA | Fold change | P value |
| ANP32A-005 | 4.08 | 0.008 | ATP5C1-006 | 0.16 | 0.033 |
| TBC1D5-021 | 3.71 | 0.001 | TMCC2-004 | 0.17 | 0.009 |
| TULP4-005 | 3.36 | 0.005 | PDCD4-010 | 0.23 | 0.045 |
| MSN-006 | 3.29 | 0.047 | SNHG7-003 | 0.27 | 0.028 |
| GDI2-006 | 3.04 | 0.029 | RAD51C-009 | 0.28 | 0.000 |
| HIST4H4-004 | 2.89 | 0.034 | ANP32A-007 | 0.32 | 0.030 |
| POLR2J3-007 | 2.87 | 0.037 | C17orf62-032 | 0.33 | 0.006 |
| PMS2P3-004 | 2.77 | 0.042 | SGK1-022 | 0.34 | 0.021 |
| C9orf78-006 | 2.57 | 0.000 | CTD-2561J22.5-004 | 0.34 | 0.013 |
| SCAF11-011 | 2.51 | 0.016 | RP11-820I16.4-001 | 0.36 | 0.003 |
| SERINC3-003 | 2.46 | 0.009 | DDX11L1-002 | 0.40 | 0.032 |
| VCL-007 | 2.42 | 0.018 | PPIF-005 | 0.40 | 0.034 |
| AC093323.3-002 | 2.38 | 0.007 | CROCCP3-006 | 0.40 | 0.007 |
| VPS39-004 | 2.37 | 0.009 | RP11-185E8.1-001 | 0.41 | 0.004 |
| AC005154.6-015 | 2.36 | 0.033 | CHIT1-004 | 0.44 | 0.017 |
| MORC3-010 | 2.35 | 0.001 | DCAF17-002 | 0.44 | 0.018 |
| LINC01128-004 | 2.33 | 0.018 | CTD-3203P2.3-001 | 0.45 | 0.035 |
| CANX-016 | 2.33 | 0.018 | GS1-124K5.3-001 | 0.46 | 0.047 |
| GS1-124K5.3-002 | 2.32 | 0.004 | GS1-279B7.2-001 | 0.47 | 0.020 |
| ZFAND5-007 | 2.26 | 0.011 | FAM228B-002 | 0.47 | 0.000 |
| STAG3L5P-002 | 2.26 | 0.023 | RBM39-027 | 0.47 | 0.023 |
| KLHL20-003 | 2.22 | 0.005 | GDAP2-003 | 0.48 | 0.009 |
| PAPSS1-008 | 2.20 | 0.039 | DUSP22-010 | 0.48 | 0.034 |
| LEPROT-005 | 2.16 | 0.041 | PPP1R37-006 | 0.48 | 0.001 |
| FHL3-005 | 2.15 | 0.008 | CNDP2-030 | 0.48 | 0.001 |
| RBM17-011 | 2.15 | 0.018 | LINC01151-001 | 0.48 | 0.014 |
| PRR5L-013 | 2.15 | 0.011 | |||
| EIF4H-005 | 2.14 | 0.016 | |||
| ERICH1-006 | 2.13 | 0.005 | |||
| ARF1-011 | 2.09 | 0.034 | |||
| ADARB1-001 | 2.09 | 0.004 | |||
| FXR2-006 | 2.08 | 0.009 | |||
| BRD7-003 | 2.08 | 0.021 | |||
| MYLK-016 | 2.07 | 0.001 | |||
| VRK3-012 | 2.02 | 0.029 | |||
| RP11-20D14.6-001 | 2.01 | 0.047 | |||
Transcripts of lncRNAs with fold change (unstable/stable) ≥ 2 and P value < 0.05 were considered as differentially expressed transcripts. n = 3 for each group.
Verification of selected candidate lncRNAs by comparing their expression levels in blood samples by qPCR
The expression levels of candidate lncRNAs (ANP32A-005, TULP4-005, PDCD4-010, and SNHG7-003) in 30 blood samples (15 patients in the stable plaque group and 15 patients in the unstable plaque group) were determined using RT-qPCR. These patients whose peripheral blood samples were used for lncRNAs validation were age and sex matched, and have largely comparable incidence of hypertension and diabetes mellitus, as well as smoking ratio (Table 1), which potentially excluded the interference of other possible factors on lncRNA expression. As shown in Figure 3A, the expression level of lncRNA-SNHG7-003 was significantly lower in blood samples of patients in the unstable plaque group, which was consistent with the RNA-Seq results (Figure 3B). The expression levels of ANP32A-005 and PDCD4-010 were not significantly different between the unstable plaque group and the stable plaque group. The expression level of lncRNA-TULP4-005 in the unstable plaque group was also lower. However, this was opposite to the RNA-Seq results, which showed an up-regulated expression of lncRNA-TULP4-005 in patients with unstable plaques (Figure 3B). The contradictions between our qPCR and RNA-Seq assays were probably due to the big variation in the RNA-Seq assay with a much smaller sample size. Therefore, the candidate lncRNA-SNHG7-003 was validated to be the lncRNA with differentially down-regulated expression in the blood of CAD patients with unstable plaque.
Figure 3.

Comparison on the expression of candidate lncRNAs in the unstable plaque and stable plaque groups. A. The expression levels of candidate lncRNA were measured in a validation cohort of patients by RT-qPCR. n = 15 for each group. B. The expression levels of candidate lncRNAs were retrieved from the RNA-Seq data. n = 3 for each group. FPKM, Fragments Per Kilobase of transcript per Million mapped reads. *P < 0.05.
lncRNA-SNHG7-003 was down-regulated in LPS induced inflammatory cell models
Considering the critical roles of inflammation in the pathogenesis of atherosclerosis [24-26], we examined the expression levels of candidate lncRNAs (ANP32A-005, TULP4-005, PDCD4-010, and SNHG7-003) in the human monocytic cell line THP-1 cells. LPS stimulation was employed to mimic the inflammatory signal in THP-1 monocytes and THP-1-derived macrophages. Compared with the THP-1 cells without LPS treatment, lncRNA-SNHG7-003 was significantly down-regulated in LPS stimulated THP-1 monocytes (Figure 4A) and macrophages (Figure 4B), while the expression of ANP32A-005, TULP4-005, and PDCD4-010 showed no difference between the inflammatory cells and control cells.
Figure 4.
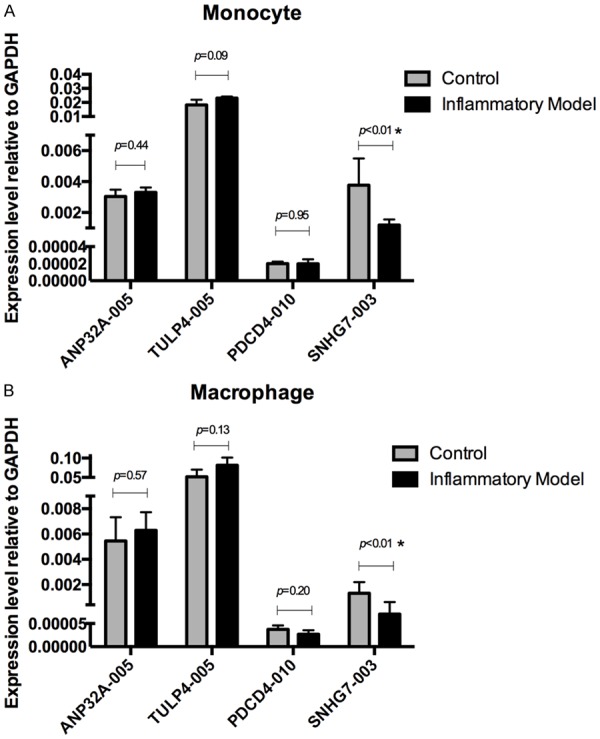
Comparison on the expression levels of candidate lncRNAs in LPS induced inflammatory cell models. (A, B) The expression levels of indicated lncRNAs were quantitated by RT-qPCR in THP-1 monocytes (A) and THP-1-derived macrophages (B) without LPS treatment (Control) or with stimulation by 1 μg/ml LPS for 24 hours (Inflammatory Model). n = 3 for each group; *P < 0.01. Data represent one of three experiments with similar results.
Activation of NF-κB in LPS stimulated monocytes and macrophages was inhibited by over-expression of lncRNA-SNHG7-003
To further elucidate the molecular mechanisms of lncRNA-SNHG7-003 in regulating inflammation, we explored the impacts of over-expression of lncRNA-SNHG7-003 on inflammatory responses of THP-1 cells. First, we determined the overexpression efficiency of lncRNA-SNHG7-003 by RT-qPCR analysis. As shown in Figure 5, the expression levels of SNHG7-003 in THP-1 monocytes or macrophages transfected with pcDNA3.1+/SNHG7-003 recombinant plasmid were much higher than those transfected with pcDNA3.1+ control plasmid, which indicated that SNHG7-003 was successfully overexpressed in THP-1 monocytes and macrophages.
Figure 5.

Verification of SNHG7-003 over-expression in THP-1 cells. The expression levels of SNHG7-003 in THP-1 monocytes or THP-1-derived macrophages transfected with pcDNA3.1+/SNHG7-003 recombinant plasmid or pcDNA3.1+ control plasmid were quantitated by RT-qPCR. n = 3 for each group; *P < 0.001. Data represent one of three experiments with similar results.
Next, we evaluated the effects of SNHG7-003 over-expression on NF-κB activation in LPS induced THP-1 monocytes and macrophages by western blot analysis. As shown in Figure 6, the levels of phosphorylated NF-κB p65 (S5-36), a hallmark of NF-κB activation, were significantly reduced in LPS stimulated THP-1 monocytes and macrophages when lncRNA-SNHG7-003 was over-expressed. On the contrary, over-expression of SNHG7-003 didn’t show effects on NF-κB activation in THP-1 monocytes and macrophages without LPS stimulation. Moreover, the expression levels of basal NF-κB p65 in THP-1 monocytes and macrophages were not influenced by either SNHG7-003 over-expression or LPS stimulation (Figure 6).
Figure 6.

Activation of NF-κB in LPS stimulated monocytes and macrophages was inhibited by over-expression of lncRNA-SNHG7-003. A. Representative western blot results showing effects of SNHG7-003 over-expression on NF-κB activation in LPS stimulated THP-1 monocytes and THP-1-derived macrophages. B. Summarized results on the relative expression levels of phosphorylated NF-κB (p65). n = 3 for each group; *P < 0.05. Data represent one of three experiments with similar results.
Inflammatory mediators were reduced by over-expression of lncRNA-SNHG7-003 in LPS stimulated monocytes and macrophages
We also evaluated the effects of SNHG7-003 over-expression on the production of inflammatory mediators in LPS stimulated THP-1 monocytes and macrophages by ELISA. These soluble inflammatory factors included TNF-α, IL-1β, MCP-1 and MMP-9, which are considered as major driving forces of atherosclerotic lesion development [38,39]. As shown in Figure 7, LncRNA-SNHG7-003 over-expression in both monocytes and macrophages significantly reduced the secretion of LPS induced inflammatory mediators (TNF-α, IL-1β, MCP-1 and MMP-9) to cell culture supernatants. However, over-expression of SNHG7-003 didn’t show significant effects on the production of these inflammatory factors in THP-1 monocytes and macrophages without LPS stimulation (Figure 7).
Figure 7.

ELISA results showing the effects of SNHG7-003 over-expression on the production of inflammatory mediators in THP-1 cells. The levels of soluble inflammatory factors, including TNF-α, IL-1β, MCP-1 and MMP-9, secreted from THP-1 monocytes and THP-1-derived macrophages with or without LPS stimulation were quantiated by ELISA. n = 3 for each group; *P < 0.05, **P < 0.01. Data represent one of at least three experiments with similar results.
Discussion
Cardiovascular disease (CVD) is the leading cause of global death [40], and ACS is one of the most life-threatening CVDs, with an estimated mortality rate of one third worldwide [41]. Early and timely recognition of unstable plaques would help with effective intervention in patients with CAD, and thus significantly improve their prognosis. In this study, we profiled lncRNAs in blood samples from CAD patients with unstable and stable plaques by RNA-Seq, and identified a series of lncRNAs that were differentially expressed in peripheral blood of patients with unstable plaques. LncRNA-SNHG7-003 was validated to be significantly down-regulated in blood samples of patients with unstable plaques and LPS-stimulated THP-1 monocytes and THP-1-derived macrophages. Over-expression of SNHG7-003 markedly inhibited the activation of NF-κB pathway, and reduced the secretion of inflammatory mediators (TNF-α, IL-1β, MCP-1 and MMP-9) in LPS-activated THP-1 monocytes and macrophages. Our work suggests that LncRNA-SNHG7-003 is a potential blood biomarker for evaluating plaque stability.
Increasing studies have demonstrated that many circulation biomarkers could reflect inflammatory reaction and oxidative stress during atherogenesis, such as C-reactive protein, fibrinogen, interleukin-6, and interleukin-18 [42]. Additionally, molecules promoting the development and rupture of unstable plaques, such as matrix metalloproteinases (MMPs, especially MMP-9), myeloperoxidase (MPO) and MCP-1, also showed uncertain sensitivity and specificity in predicting cardiac events [42]. In recent years, some microRNAs were found to be related to plaque stability, such as miR-21, miR-100, miR-127, miR-133, miR-143/145, miR-221/222, and miR-494 [43]. Considering the pivotal role of inflammatory reaction on unstable plaques formation and rupture, many studies evaluated their predictive value for cardiac events among CAD patients. However, the results were controversial and their predictive values were far from comprehensive [42]. Therefore, effective biomarkers for unstable plaque, like troponin for myocardial infarction and BNP for heart failure, are still absent.
With the springing up of research in lncRNAs, the roles of lncRNAs in the process of atherosclerosis are gradually uncovered. Some studies have reported that several lnc-RNAs, such as OTTHUMT00000387022, ENST00000512246.1, AC100865.1, H19 and LIPCAR, might be potential diagnostic markers of coronary artery diseases [44-47]. However, lncRNAs related to coronary artery atherosclerotic plaque stability have never been identified so far. Our results showed that SNHG7-003 was significantly down-regulated in blood samples of patients with unstable plaque comparing with those with stable plaque, which was in accordance with the RNA-Seq results. Inconsistent results were observed between RTqPCR verification and initial RNA-Seq screening for other 3 candidate lncRNAs, including ANP32A-005, PDCD4-010 and TULP4-005, and this was probably due to the small sample size and large variations. Due to the technical limitation on design for specific and effective primers, we only selected 4 candidates among the very top differentially expressed lncRNAs for validation. With more improvements on primer design and larger sample sizes, more potential candidates might be validated to be clarified for their correlation with coronary artery atherosclerotic plaque stability. Nevertheless, we could, at least so far, conclude that lncRNA-SNHG7-003 is a potential circulating biomarker for evaluating plaque stability.
Atherosclerosis is a chronic inflammatory disease occurs at the arterial wall [48]. Inflammatory reaction in the plaques is the key factor leading to formation and rupture of unstable plaques [1,49]. Monocytes and monocytes-derived macrophages are two major cells involved in plaque inflammation, and play important roles in occurrence and development of unstable plaques [1,50]. In this study, the samples we used for RNA-Seq screening and RT-qPCR validations were from peripheral blood, in which monocytes are included. Acquisition of RNA samples from total peripheral blood is more convenient, which could facilitate the fast diagnosis of unstable plaques without cell purification. However, purified monocytes could be a more ideal population for studying the association between lncRNAs and inflammation. Therefore, we compared the expression levels of candidate lncRNAs in LPS stimulated THP-1 monocytes and THP-1-derived macrophages to figure out their relationship with atherosclerosis-related inflammation. Interestingly, lncRNA-SNHG7003 was also significantly down-regulated in these LPS activated monocytes and macrophages, which indicated that SNHG7-003 was potentially involved in the inflammatory reactions in monocytes and macrophages.
LPS could activate THP-1 monocytes and macrophages through Toll-like receptor 4 (TLR4) and its downstream pathways (such as NF-κB pathway), which leads to the expression and release of inflammatory factors (such as TNF-α, IL-1β and MCP-1) and matrix-degrading enzymes (such as MMP-9), and thus facilitates inflammation [51-53]. These inflammatory factors and matrix-degrading enzymes have been shown to contribute to unstable plaques formation and rupture during atherosclerosis [48,49,54,55]. Therefore, in order to validate our speculation on the role of SNHG7-003 in atherosclerosis-related inflammation, we determined NF-κB activation and expression levels of inflammatory mediators in monocytes and macrophages with SNHG7-003 over-expression. Our results showed that activation of NF-κB and expression of inflammatory mediators (TNF-α, IL-1β, MCP-1 and MMP-9) were inhibited by over-expression of SNHG7-003 in LPS stimulated monocytes and macrophages. Recently, lncRNA-SNHG7 has been reported to promote the proliferation of lung cancer cells, glioblastoma cells and colorectal cancer through multiple mechanisms [56-58]. Therefore, the reduced production of inflammatory mediators was not likely due to the decreased proliferation of LPS-stumulated monocytes and macrophages. Taking all these into account, we could at least conclude that lncRNA-SNHG7-003 can substantially weaken the inflammatory responses of LPS activated monocyte and macrophage, although the underlying molecular mechanisms still remain to be fully elucidated.
The current investigation has several strengths and limitations. First, to the best of our knowledge, this is the first study on identifying circulation lncRNAs related to coronary atherosclerotic plaque stability. Moreover, we were the first to link lncRNA-SNHG7-003 and plaque stability. Furthermore, we clarified a novel role of SNHG7-003 in LPS-activated monocytes and macrophages. However, there are some limitations in our study. First, blood samples used for RNA-Seq screening and candidate lncRNAs verification were limited and the variations were large. Further verifications are required to support the clinical use of SNHG7-003 for plaque stability evaluation. In addition, the specificity and sensitivity of SNHG7-003 in diagnosing unstable plaques were not investigated, which should be addressed in future studies.
In conclusion, we provided transcriptome-wide overview of aberrantly expressed lncRNAs in CAD patients with unstable and stable plaques, and identified SNHG7-003 as a novel lncRNA biomarker for diagnosing unstable plaques. SNHG7-003 could inhibit LPS-induced activation of NF-κB pathway, and regulate inflammatory response in human monocytes and macrophages. Blood lncRNA-SNHG7-003 has the potential to be a circulation lncRNA biomarker for evaluating plaque stability in patients with CAD.
Acknowledgements
This study was supported by grants from Shanghai Jiao Tong University School of Medicine (20172028), and the National Natural Science Foundation of China (81370400).
Disclosure of conflict of interest
None.
References
- 1.Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114:1852–1866. doi: 10.1161/CIRCRESAHA.114.302721. [DOI] [PubMed] [Google Scholar]
- 2.Narula J, Strauss HW. The popcorn plaques. Nat Med. 2007;13:532–534. doi: 10.1038/nm0507-532. [DOI] [PubMed] [Google Scholar]
- 3.Sanz J, Fayad ZA. Imaging of atherosclerotic cardiovascular disease. Nature. 2008;451:953–957. doi: 10.1038/nature06803. [DOI] [PubMed] [Google Scholar]
- 4.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2000;20:1262–1275. doi: 10.1161/01.atv.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 5.Osborn EA, Jaffer FA. Imaging atherosclerosis and risk of plaque rupture. Curr Atheroscler Rep. 2013;15:359. doi: 10.1007/s11883-013-0359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okura H, Asawa K, Kubo T, Taguchi H, Toda I, Yoshiyama M, Yoshikawa J, Yoshida K. Incidence and predictors of plaque rupture in the peripheral arteries. Circ Cardiovasc Interv. 2010;3:63–70. doi: 10.1161/CIRCINTERVENTIONS.109.900779. [DOI] [PubMed] [Google Scholar]
- 7.Okura H, Kobayashi Y, Sumitsuji S, Terashima M, Kataoka T, Masutani M, Ohyanagi M, Shimada K, Taguchi H, Yasuga Y, Takeda Y, Ohashi Y, Awano K, Fujii K, Mintz GS. Effect of culprit-lesion remodeling versus plaque rupture on three-year outcome in patients with acute coronary syndrome. Am J Cardiol. 2009;103:791–795. doi: 10.1016/j.amjcard.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Niccoli G, Montone RA, Di Vito L, Gramegna M, Refaat H, Scalone G, Leone AM, Trani C, Burzotta F, Porto I, Aurigemma C, Prati F, Crea F. Plaque rupture and intact fibrous cap assessed by optical coherence tomography portend different outcomes in patients with acute coronary syndrome. Eur Heart J. 2015;36:1377–1384. doi: 10.1093/eurheartj/ehv029. [DOI] [PubMed] [Google Scholar]
- 9.Kotani J, Mintz GS, Castagna MT, Pinnow E, Berzingi CO, Bui AB, Pichard AD, Satler LF, Suddath WO, Waksman R, Laird JR Jr, Kent KM, Weissman NJ. Intravascular ultrasound analysis of infarct-related and non-infarct-related arteries in patients who presented with an acute myocardial infarction. Circulation. 2003;107:2889–2893. doi: 10.1161/01.CIR.0000072768.80031.74. [DOI] [PubMed] [Google Scholar]
- 10.Akosah KO, Schaper A, Cogbill C, Schoenfeld P. Preventing myocardial infarction in the young adult in the first place: how do the national cholesterol education panel III guidelines perform? J Am Coll Cardiol. 2003;41:1475–1479. doi: 10.1016/s0735-1097(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Garcia HM, Jang IK, Serruys PW, Kovacic JC, Narula J, Fayad ZA. Imaging plaques to predict and better manage patients with acute coronary events. Circ Res. 2014;114:1904–1917. doi: 10.1161/CIRCRESAHA.114.302745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii K, Hao H, Ohyanagi M, Masuyama T. Intracoronary imaging for detecting vulnerable plaque. Circ J. 2013;77:588–595. doi: 10.1253/circj.cj-12-1599. [DOI] [PubMed] [Google Scholar]
- 13.Jacob SS, Hassan M, Yacoub MH. Utility of mass spectrometry for the diagnosis of the unstable coronary plaque. Glob Cardiol Sci Pract. 2015;2015:25. doi: 10.5339/gcsp.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 15.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 16.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 19.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, Tian W. Molecular mechanisms and function prediction of long noncoding RNA. ScientificWorldJournal. 2012;2012:541786. doi: 10.1100/2012/541786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 23.Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–12382. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viola J, Soehnlein O. Atherosclerosis - a matter of unresolved inflammation. Semin Immunol. 2015;27:184–193. doi: 10.1016/j.smim.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 26.Fava C, Montagnana M. Atherosclerosis is an inflammatory disease which lacks a common anti-inflammatory therapy: how human genetics can help to this issue. A narrative review. Front Pharmacol. 2018;9:55. doi: 10.3389/fphar.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 29.Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 31.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhou T, Ding JW, Wang XA, Zheng XX. Long noncoding RNAs and atherosclerosis. Atherosclerosis. 2016;248:51–61. doi: 10.1016/j.atherosclerosis.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 34.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge J, Chirillo F, Schwedtmann J, Gorge G, Haude M, Baumgart D, Shah V, von Birgelen C, Sack S, Boudoulas H, Erbel R. Screening of ruptured plaques in patients with coronary artery disease by intravascular ultrasound. Heart. 1999;81:621–627. doi: 10.1136/hrt.81.6.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran M, Chen B, Li Z, Wu M, Liu X, He C, Zhang S. Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol Reprod. 2016;94:77. doi: 10.1095/biolreprod.115.136911. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Ramji DP, Davies TS. Cytokines in atherosclerosis: key players in all stages of disease and promising therapeutic targets. Cytokine Growth Factor Rev. 2015;26:673–685. doi: 10.1016/j.cytogfr.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprague AH, Khalil RA. Inflammatory cytokines in vascular dysfunction and vascular disease. Biochem Pharmacol. 2009;78:539–552. doi: 10.1016/j.bcp.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laslett LJ, Alagona P Jr, Clark BA 3rd, Drozda JP Jr, Saldivar F, Wilson SR, Poe C, Hart M. The worldwide environment of cardiovascular disease: prevalence, diagnosis, therapy, and policy issues: a report from the American college of cardiology. J Am Coll Cardiol. 2012;60:S1–49. doi: 10.1016/j.jacc.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Grech ED, Ramsdale DR. Acute coronary syndrome: unstable angina and non-ST segment elevation myocardial infarction. BMJ. 2003;326:1259–1261. doi: 10.1136/bmj.326.7401.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah PK. Biomarkers of plaque instability. Curr Cardiol Rep. 2014;16:547. doi: 10.1007/s11886-014-0547-7. [DOI] [PubMed] [Google Scholar]
- 43.Koroleva IA, Nazarenko MS, Kucher AN. Role of microRNA in development of instability of atherosclerotic plaques. Biochemistry (Mosc) 2017;82:1380–1390. doi: 10.1134/S0006297917110165. [DOI] [PubMed] [Google Scholar]
- 44.Cai Y, Yang Y, Chen X, Wu G, Zhang X, Liu Y, Yu J, Wang X, Fu J, Li C, Jose PA, Zeng C, Zhou L. Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res. 2016;112:714–724. doi: 10.1093/cvr/cvw022. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Zhao Z, Gao C, Rao L, Hao P, Jian D, Li W, Tang H, Li M. Identification of a peripheral blood long non-coding RNA (upperhand) as a potential diagnostic marker of coronary artery disease. Cardiol J. 2018;25:393–402. doi: 10.5603/CJ.a2017.0133. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Cai Y, Wu G, Chen X, Liu Y, Wang X, Yu J, Li C, Jose PA, Zhou L, Zeng C. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci (Lond) 2015;129:675–685. doi: 10.1042/CS20150121. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Gao W, Long QQ, Zhang J, Li YF, Liu DC, Yan JJ, Yang ZJ, Wang LS. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7:7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansson GK, Libby P, Tabas I. Inflammation and plaque vulnerability. J Intern Med. 2015;278:483–493. doi: 10.1111/joim.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah PK. Molecular mechanisms of plaque instability. Curr Opin Lipidol. 2007;18:492–499. doi: 10.1097/MOL.0b013e3282efa326. [DOI] [PubMed] [Google Scholar]
- 50.Hilgendorf I, Swirski FK, Robbins CS. Monocyte fate in atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35:272–279. doi: 10.1161/ATVBAHA.114.303565. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Murshid A, Borges TJ, Lang BJ, Calderwood SK. The scavenger receptor SREC-I cooperates with toll-like receptors to trigger inflammatory innate immune responses. Front Immunol. 2016;7:226. doi: 10.3389/fimmu.2016.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edfeldt K, Swedenborg J, Hansson GK, Yan ZQ. Expression of toll-like receptors in human atherosclerotic lesions: a possible pathway for plaque activation. Circulation. 2002;105:1158–1161. [PubMed] [Google Scholar]
- 54.Gopalakrishnan M, Silva-Palacios F, Taytawat P, Pant R, Klein L. Role of inflammatory mediators in the pathogenesis of plaque rupture. J Invasive Cardiol. 2014;26:484–492. [PubMed] [Google Scholar]
- 55.Diez J. Emerging role of matrix metalloproteinases in the pathophysiology of cardiac diseases. Eur J Clin Invest. 2002;32:291–294. doi: 10.1046/j.1365-2362.2002.00980.x. [DOI] [PubMed] [Google Scholar]
- 56.Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, Sun Q. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496:712–718. doi: 10.1016/j.bbrc.2018.01.109. [DOI] [PubMed] [Google Scholar]
- 57.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 58.Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9:722. doi: 10.1038/s41419-018-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


