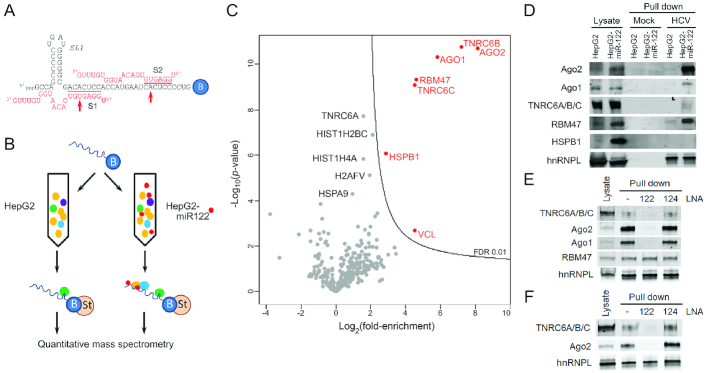
Figure 1.
TNRC6 proteins bind to HCV RNA in a miR-122-dependent fashion. (A) RNA bait used in pull-down reactions, showing base-pair interactions between miR-122 and the HCV 5′ UTR terminus. Seed sequence interactions at S1 and S2 are highlighted. Red arrows denote positions of p6m mutants. B = 3′ biotin tag. (B). Experimental scheme: the 3′ biotinylated RNA bait was bound to streptavidin beads and incubated with HepG2 or HepG2-miR-122 lysates. Proteins associated with the RNA bait were isolated and subjected to quantitative mass spectrometry. (C) Volcano plot of quantitative mass spectrometry results showing the 324 proteins that were confidently identified in (B) plotted according to their degree of enrichment in the pull-down products from the HepG2-miR122 versus miR-122-deficident HepG2 lysate: log2 (fold-enrichment) versus probability, -log10(P-value). The contour line indicates the enrichment FDR of 0.01. Proteins enriched with an FDR < 0.01 are labeled in red, whereas other proteins of potential interest are labeled in black. (D) Immunoblot confirmation of mass spectrometry results. Protein samples from lysates, mock pull-down (beads only) and RNA bait pull-downs were resolved by SDS-PAGE and blotted with indicated antibodies. TNRC6 proteins were detected with a pan-paralog antibody recognizing TNRC6A, 6B and 6C. hnRNPL binds RNA in a miR-122-independent fashion and was included as a control. (E) Pull-down assays were carried out as in (D) in HepG2-miR-122 lysate with or without prior addition of miR-122 LNA or control miR-124 LNA antagomirs. The anti-miR-122 oligo blocked the pull-down of Ago2, Ago1 and TNRC6 proteins but not RBM47. (F) Pull-down assays as in (E) were carried out with Huh-7.5 cell lysate.