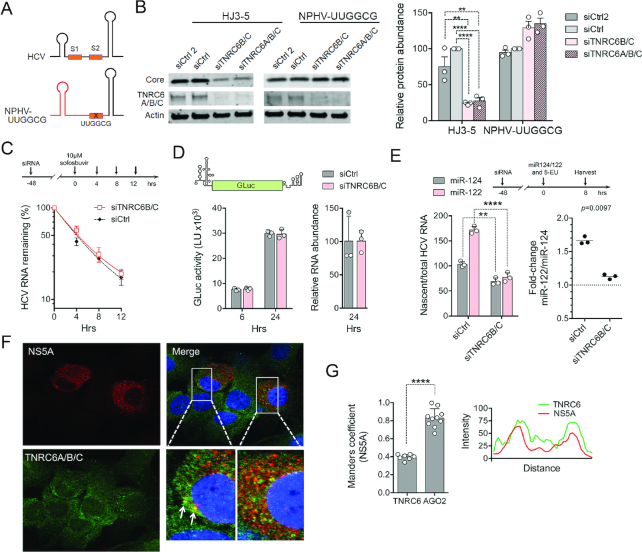
Figure 3.
TNRC6 promotes miR-122 augmented viral RNA synthesis but not HCV RNA stability or IRES activity. (A) Schematic representation of the 5′ terminal sequences of HCV and NPHV-UUGGCG viruses, with sequence from NPHV shown in red. (B) Huh7.5 cells were transfected with siCtrl (a pool of four nontargeting siRNAs), siCtrl2 (an additional single nontargeting siRNA) or the indicated TNRC6-targeting siRNAs, and 48 h later re-transfected with viral RNA. Viral and cellular protein abundance was analyzed by immunoblot after an additional 72 h. HCV core protein abundance was quantified relative to actin abundance in three independent experiments, with results shown on the right as means ± s.d. after normalizing to the siCtrl control. ****adjusted P-value < 0.0001, **adjusted P-value 0.0011–0.0019 by two-way ANOVA with Sidak’s test for multiple comparisons. (C) Positive-strand HCV RNA stability in cells following sofosbuvir arrest of viral RNA synthesis. HCV RNA level at 0 h was arbitrarily set to 100%. Data are means from three independent experiments ± s.e.m. Weighted R2 = 0.975–0.987 when fit to a single-phase decay model, with no significant difference in the decay rate, k, by extra sum-of-squares F test (P > 0.05). (D) HCV IRES activity. TNRC6-depleted Huh-7.5 cells were transfected with HCV mini-genome RNA containing GLuc sequence flanked by viral 5′ and 3′ UTRs. (Left) GLuc activities were measured in supernatant fluids, and (right) relative reporter RNA abundance determined in cell lysates (siCtrl cells arbitrarily set to 100). Data are means from two independent experiments, one with two technical replicates (shaded symbols) ± s.d. (E) Nascent HCV RNA synthesis in TNRC6-depleted cells. (Top) Stably infected Huh-7.5 cells were transfected with indicated siRNAs, and 48 h later re-transfected with miR-122, or the control miR-124, and 5-ethynyluridine (5-EU) added to the medium. Cells were harvested 8 h later for the isolation of nascent RNA. (Bottom left) The relative rate of viral RNA synthesis was calculated as nascent HCV RNA divided by total HCV RNA, with viral RNA synthesis in siCtrl control cells transfected with miR-124 arbitrarily set to 100. **P < 0.01, ****P < 0.0001 by two-way ANOVA with Sidak’s multiple comparison test. (Bottom right) miR-122-induced fold-change in viral RNA synthesis relative to that in control cells transfected with miR124. P = 0.0097 by two-sided paired t-test. Data are means of three independent experiments ± s.d. (F) Confocal fluorescence microscopy of Huh-7.5 cells infected with HJ3-5 virus and stained with antibodies to NS5A (red) and TNRC6 (green). Nuclei counterstained with DAPI (blue) in the merged image. Arrows denote puncta with co-localization of NS5A and TNRC6 proteins. (G) Quantitative analysis of confocal microscopy images. (Left) Manders’ coefficient for co-localization of TNRC6 proteins and NS5A and, for comparison, Ago2 and NS5A in similar images (see Supplementary Figure S4). ****P < 0.0001 by two-sided t-test, n = 7–10 cells. (Right) Linear trace of TNRC6 and NS5A fluorescent intensities bisecting the two puncta depicted by arrows in panel (F).