Figure 1.
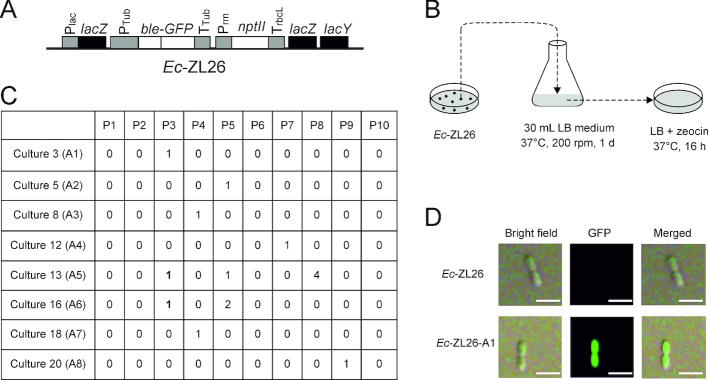
Genetic screen for functional activation of a eukaryotic gene following horizontal transfer into a prokaryote (Escherichia coli). (A) Schematic map of the eukaryotic ble-GFP gene transferred to the lacZ locus in the E. coli genome by selection for kanamycin resistance (conferred by the nptII gene). The ble-GFP fusion gene is driven by the tubulin gene promoter (PTub) and terminator (TTub) from the red alga Porphyridium purpureum (35) and confers resistance to the antibiotic zeocin. Ec-ZL26 denotes the E. coli strain harboring the construct in its lacZ locus. Plac, lac operon promoter; Prrn, rRNA operon promoter from the tobacco plastid genome; TrbcL, 3′ UTR from the tobacco rbcL gene. Endogenous bacterial genes are represented as black boxes, coding regions of transgenes as white boxes and promoters and terminators (driving transgenes and the lac operon) as gray boxes. (B) Selection for functional activation of the ble-GFP gene. A large-scale genetic screen for functional activation of the ble gene in bacteria was conducted by exposing cultures of Ec-ZL26 to stringent selection for ble-mediated resistance to zeocin. The 30 ml LB medium inoculated with a single colony of Ec-ZL26 was cultivated for 1 day, followed by plating of equal aliquots onto 10 LB plates with zeocin for overnight selection of resistant colonies. The selection process was repeated 30 times (i.e. with 30 independent cultures of 30 ml each). In total, approximate 1012E. coli cells were subjected to selection on 300 plates. (C) Zeocin-resistant colonies appeared on selection plates from 8 of the 30 cultures. P, number of the selection plate; A, gene activation event; 1 (in bold), the colony representing the initially characterized activation event from cultures 13 and 16. (D) Analysis of GFP accumulation in a dividing zeocin-resistant cell from activation event Ec-ZL26-A1 by confocal laser-scanning microscopy. Scale bars: 1 μm.