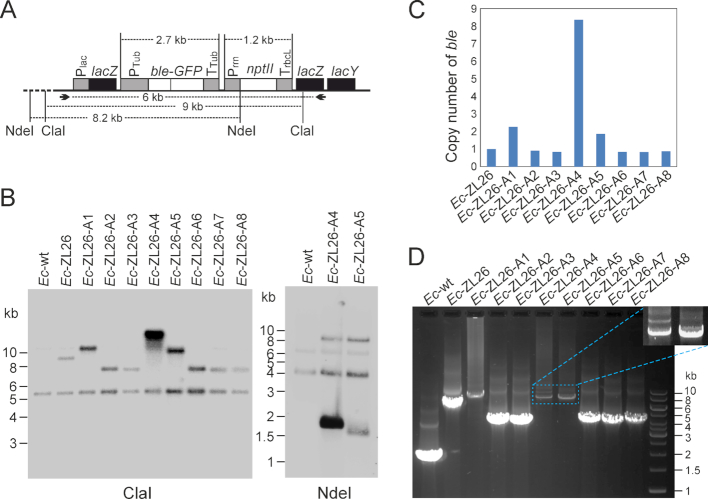
Figure 3.
Identification of genomic rearrangements leading to functional activation of the eukaryotic ble-GFP gene in Escherichia coli. (A) Map of the region transferred into the E. coli genome. Restriction sites used in RFLP analyses and primers used for PCR amplification and DNA sequencing are indicated, and expected fragment sizes are given in kb. (B) Southern blot analyses of zeocin-resistant clones. Total bacterial DNA was digested with two different restriction enzymes. While ClaI does not have a recognition site in the transferred DNA, NdeI cuts once in the transgenic region (at the start codon of the nptII coding region). The blots were hybridized to a radiolabeled probe covering the ble coding region. The sizes of the restriction fragments recognized in the Ec-ZL26 strain are 9023 bp for ClaI and 8191 bp for NdeI. (C) Quantification of the bands in the Southern blot assays (with the Image Lab 6.0 software) to estimate the copy number of the ble gene in the bacterial strains with activated ble expression. The cross-hybridizing band of ∼5.1 kb (in the ClaI blot) that is also visible in the wild type was used as reference band for quantitation. (D) Gel electrophoretic analysis of PCR products of the region harboring the activated ble (obtained with the primers indicated in panel A). The magnified area shows a stronger exposure of the amplified bands corresponding to the tandem duplications in strains A4 and A5 (cf. Figure 4).