Figure 3.
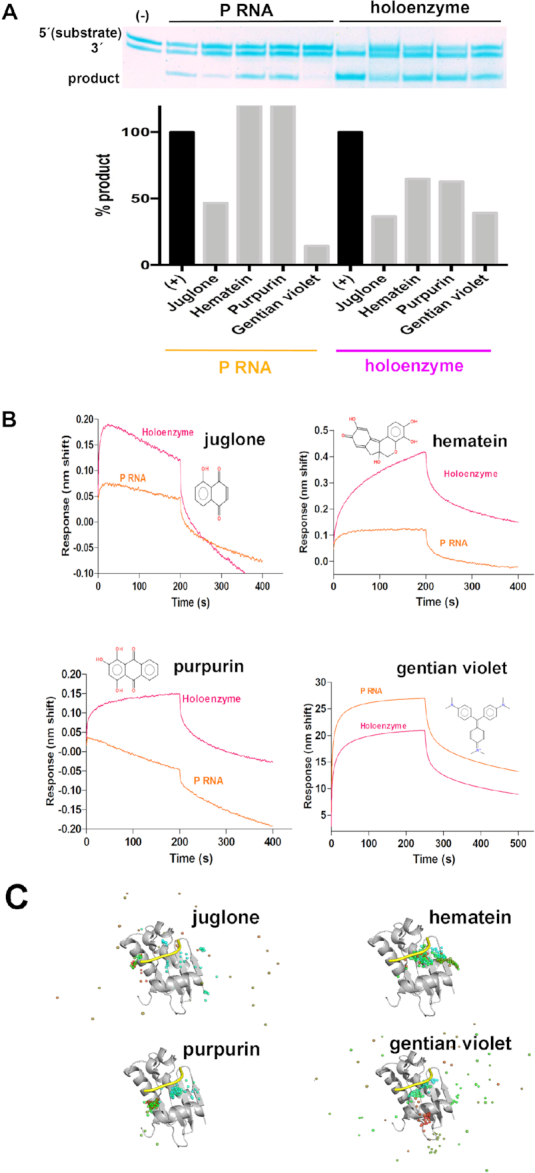
The inhibitors purpurin and hematein bind to the holoenzyme. (A) Top. 19% urea-PAGE demonstrates bipartite pre-tRNA substrate is processed with either P RNA or the RNase P holoenzyme from T. maritima. Bottom. Comparison of RNase P activity using P RNA (left) or the holoenzyme (right), indicates that purpurin and hematein do not inhibit the P RNA reaction, as derived by densitometry analysis of the lower band (product). (B) The relative affinities of the inhibitors for the P RNA or the holoenzyme differ. Binding sensorgrams obtained by biolayer interferometry (BLI). P RNA or RNase P holoenzyme was immobilized on super streptavidin (SSA) biosensors through a 5′-biotinylated RNA oligonucleotide complementary to a region on the P1 stem of P RNA (see Materials and Methods). Remarkably, purpurin does not bind to P RNA (orange). In contrast, gentian violet binds not only to P RNA but also to the Mh substrate (Supplementary Figure S16). (C) Molecular Dynamics (MD) simulations of RNase P protein in complex with the validated inhibitors. In each panel, semitransparent spheres represent the geometric center, over a 200 ns trajectory, of the corresponding tested compound. The 5′-leader (yellow) is shown for reference purposes only; it was not included in the simulation. Sphere color coding indicates the starting and final positions in blue and red, respectively. The starting complex conformation for gentian violet, juglone and hematein correspond to the best pose suggested by docking simulations. Purpurin's starting conformation corresponds to that observed in the crystal structure in complex with the P protein (PDB code: 6MAX. See Figure 5). Hematein and purpurin remain bound to the P protein throughout the dynamics protocol.
