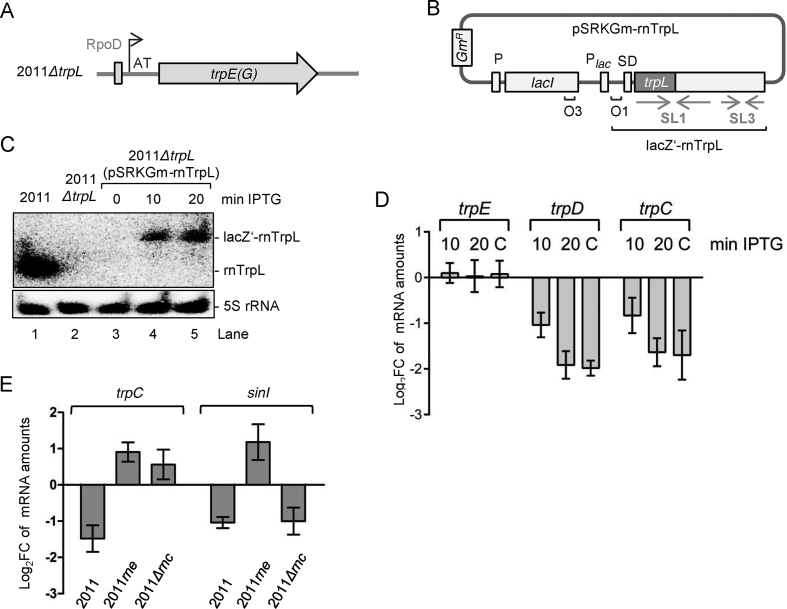
Figure 3.
Short-term overproduction of recombinant lacZ’-rnTrpL transcript in ΔtrpL background decreases the level of trpDC mRNA. (A) Schematic representation of the trpE(G) locus in the deletion mutant 2011ΔtrpL. The original TSS including the first two nucleotides (AT) was preserved. (B) Scheme of pSRKGm-rnTrpL. The pSRK-based plasmids harbor the lac repressor gene lacI with its own promoter, the lacI-lacZ intergenic region, the lac-promoter Plac, and the 38-nt lacZ leader containing the Shine-Dalgarno sequence (SD), followed by an NdeI restriction site containing the ATG translation start codon. Positions of the LacI-binding operators O1 and O3, the proper spacing of which ensures tight regulation at Plac, are indicated (42). The rnTrpL sequence was cloned in frame in the NdeI site of pSRKGm. The resulting sRNA lacZ‘-rnTrpL contains the 38-nt lacZ-leader. The scheme shows the plasmid conferring resistance to gentamycin (Gm) (pSRKGm-rnTrpL). A similar plasmid conferring resistance to tetracycline (Tc) was also constructed. (C) Northern blot hybridization showing IPTG-induced lacZ’-rnTrpL transcription at 10 and 20 min post induction in strain 2011ΔtrpL (pSRKGm-rnTrpL) (lanes 3, 4 and 5). Data for control strains are shown in lanes 1 and 2. Also shown are the hybridization data using a 5S rRNA-specific probe (loading control). (D) qRT-PCR analysis of trpD, trpC and trpE (control) expression. At 10 min and 20 min post induction with IPTG, mRNA levels of the respective mRNA in 2011ΔtrpL (pSRKGm-rnTrpL) were measured and compared to the levels prior to induction. Bar C represents the mRNA levels in the constitutively overexpressing strain 2011 (pRK-rnTrpL) compared to those of the EVC. (E) qRT-PCR analysis of trpC and the control mRNA sinI in strain 2011 and the RNase mutant strains 2011rne and 2011Δrnc (indicated). Each strain contains the plasmid pSRKTc-rnTrpL. Changes in the levels of the respective mRNA were determined 10 min post induction with IPTG. For other details, see D). The graphs show results from three independent experiments, each performed in technical duplicates (mean values and standard deviations are indicated).