Abstract
Mycobacteria and related organisms in the Corynebacterineae suborder are characterized by a distinctive outer membrane referred to as the mycomembrane. Biosynthesis of the mycomembrane occurs through an essential process called mycoloylation, which involves antigen 85 (Ag85)-catalyzed transfer of mycolic acids from the mycoloyl donor trehalose monomycolate (TMM) to acceptor carbohydrates and, in some organisms, proteins. We recently described an alkyne-modified TMM analogue (O-AlkTMM-C7) which, in conjunction with click chemistry, acted as a chemical reporter for mycoloylation in intact cells and allowed metabolic labeling of mycoloylated components of the mycomembrane. Here, we describe the synthesis and evaluation of a toolbox of TMM-based reporters bearing alkyne, azide, trans-cyclooctene, and fluorescent tags. These compounds gave further insight into the substrate tolerance of mycoloyltransferases (e.g., Ag85s) in a cellular context and they provide significantly expanded experimental versatility by allowing one- or two-step cell labeling, live cell labeling, and rapid cell labeling via tetrazine ligation. Such capabilities will facilitate research on mycomembrane composition, biosynthesis, and dynamics. Moreover, because TMM is exclusively metabolized by Corynebacterineae, the described probes may be valuable for the specific detection and cell-surface engineering of Mycobacterium tuberculosis and related pathogens. We also performed experiments to establish the dependence of probe incorporation on mycoloyltransferase activity, results from which suggested that cellular labeling is a function not only of metabolic incorporation (and likely removal) pathway(s), but also accessibility across the envelope. Thus, whole-cell labeling experiments with TMM reporters should be carefully designed and interpreted when envelope permeability may be compromised. On the other hand, this property of TMM reporters can potentially be exploited as a convenient way to probe changes in envelope integrity and permeability, facilitating drug development studies.
Keywords: Corynebacterineae, mycobacteria, mycomembrane, mycolic acid, trehalose, chemical reporter, bioorthogonal chemistry, imaging
Graphical Abstract
Modifying mycobacteria via mycoloylation: A suite of trehalose monomycolate (TMM)-based metabolic reporters provides versatility and specificity for analyzing and engineering the outer membrane of living mycobacteria.
Introduction
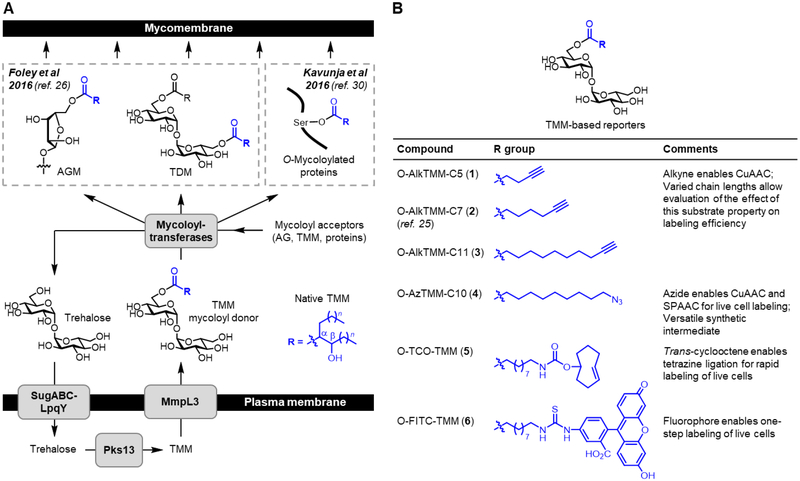
Bacteria in the Corynebacterineae suborder include species of enormous medical, biotechnological, and environmental importance, such as the global pathogen Mycobacterium tuberculosis (Mtb)[1,2] and the industrial fermentation workhorse Corynebacterium glutamicum (Cglut).[3] These bacteria have a complex cell envelope featuring a distinctive outer membrane—called the mycomembrane—which consists mainly of long-chain, branched mycolic acids that are conjugated to carbohydrates and other molecules (Figure 1A).[4–7] The mycomembrane is essential for survival due to its roles in fundamental physiological processes such as defense, nutrient acquisition, and cell–cell communication, including host–pathogen interactions.[6,7] As a result, there is significant interest in improving our understanding of mycomembrane composition, biosynthesis, and function, as this information is crucial for activities such as tuberculosis drug and diagnostic development.[8] However, progress has been hampered by the difficulty of analyzing mycomembrane biosynthetic pathways and components using traditional microbiological and biochemical techniques (e.g., radiolabeling and fractionation), particularly in native settings.[9,10]
Figure 1.
(A) TMM is the mycoloyl donor used to construct the major mycomembrane components, including AGM, TDM, and O-mycoloylated proteins. O-AlkTMM-C7 (2) is a previously developed TMM-mimicking reporter molecule that allows metabolic labeling and analysis of mycomembrane components in intact cells. (B) Chemical structures and properties of the expanded set of TMM-based reporters (1–6) developed in this work.
Metabolic reporters that exploit endogenous biosynthetic pathways are valuable tools for studying cell envelope components in intact bacterial cells.[11] Such efforts in mycobacteria have benefited from the recent elucidation of key steps in mycomembrane biosynthesis, which are mediated by the non-mammalian disaccharide trehalose.[12] Trehalose shuttles mycolic acids from the cytoplasm to the cell’s exterior to assemble the mycomembrane (Figure 1A). Trehalose is first converted to trehalose monomycolate (TMM),[13] which, after transport across the plasma membrane,[14] donates its mycoloyl group to acceptor molecules.[15] This process, termed mycoloylation, is catalyzed by a group of mycoloyltransferases, which in mycobacterial species are collectively referred to as the antigen 85 (Ag85) complex.[15–17] The major products of mycoloylation are arabinogalactan-linked mycolate (AGM) and trehalose dimycolate (TDM), which are the principal constituents of the inner- and outer-leaflets of the mycomembrane, respectively.[7] In some members of the Corynebacterineae, including Cglut, mycomembrane proteins can also be O-mycoloylated on serine residues,[18] although the scope and functional importance of this unique post-translational modification remains largely unknown.
Trehalose metabolic pathways in mycobacteria have been successfully exploited for the purpose of probe development. Fluorescein- and azide-modified trehalose analogues can be metabolically incorporated into trehalose-containing glycolipids in the mycomembrane, i.e. TMM and/or TDM, allowing detection of these species in cells either (i) directly, when the fluorophore is pre-attached to trehalose, or (ii) after click chemistry, wherein the fluorophore is delivered in a second step.[19,20] These trehalose- based reporters have been used for various applications. For instance, fluorescein-tagged trehalose analogues have been used to image Mtb within macrophages and were recently used to investigate mycomembrane fluidity in response to antitubercular drugs.[21] In addition, 6-azido trehalose labeling, deployed in conjunction with click chemistry, was used to confirm the function of MmpL3 as the essential TMM flippase and to characterize antitubercular compounds that inhibit it.[22] Fluorine-modified trehalose analogues can also be metabolized by mycobacteria, a finding which may pave the way for imaging mycobacterial infections in vivo using positron emission tomography (PET).[23,24] Recently, a trehalose analogue bearing a solvatochromic moiety was used as a fluorogenic probe for the rapid detection of Mtb in patient sputum samples.[25] The latter two examples highlight the potential of trehalose-based metabolic reporters for tuberculosis diagnostic applications.
Recently, our group introduced a new class of reporters, which are based on TMM and provide complementary capabilities for studying the mycomembrane.[26] The reporter we initially developed, named O-AlkTMM-C7, mimics the structure of the mycoloyl donor TMM, but in place of a native branched lipid we added a linear, truncated (C7) chain containing a clickable terminal alkyne tag (2, Figure 1B). When fed to mycobacterial or corynebacterial cells, O-AlkTMM-C7 was processed by mycoloyltransferases (e.g., Ag85), thus depositing its clickable lipid chain onto mycoloyl acceptors and producing species such as alkyne-labeled AGM and TDM. By combining O-AlkTMM-C7 labeling with Cu-catalyzed azide–alkyne cycloaddition (CuAAC[27,28])-mediated fluorophore delivery, various aspects of mycomembrane composition and dynamics can be probed. For instance, O-AlkTMM-C7 allowed the first direct visualization of AGM biosynthesis during cell growth and division in M. smegmatis (Msmeg).[26] O-AlkTMM-C7 was also used to assist in the functional characterization of a protein involved in mycomembrane construction, namley the mycobacterial lipid transporter LprG.[29] In another example, O-AlkTMM-C7 was shown to label mycomembrane-resident O-mycoloylated proteins in Cglut (via the pathway shown in Figure 1A), providing a chemical biology approach to the discovery and characterization of this distinctive class of proteins.[30] In the context of studying this post-translational modification, the ability of O-AlkTMM-C7 to exclusively label O-mycoloylated proteins provides a substantial advantage over using alkynyl fatty acids, which label all types of lipidated proteins in bacteria and thus lack selectivity for O-mycoloylation.[31,32] Overall, O-AlkTMM-C7 is a powerful new tool for studying various aspects of the mycomembrane. However, it is limited in some contexts by its reliance on CuAAC—which is often not suitable for use in living systems due to copper toxicity or perturbations resulting from oxidative stress—as well as the need to perform two steps to deliver a fluorophore (or other type of functional tag) to the cell surface. In this work, we sought to further develop, characterize, and define the utility of the TMM class of reporters for investigating the mycomembrane-containing species of the Corynebacterineae suborder.
Results and Discussion
Design and Synthesis of an Expanded Set of TMM reporters
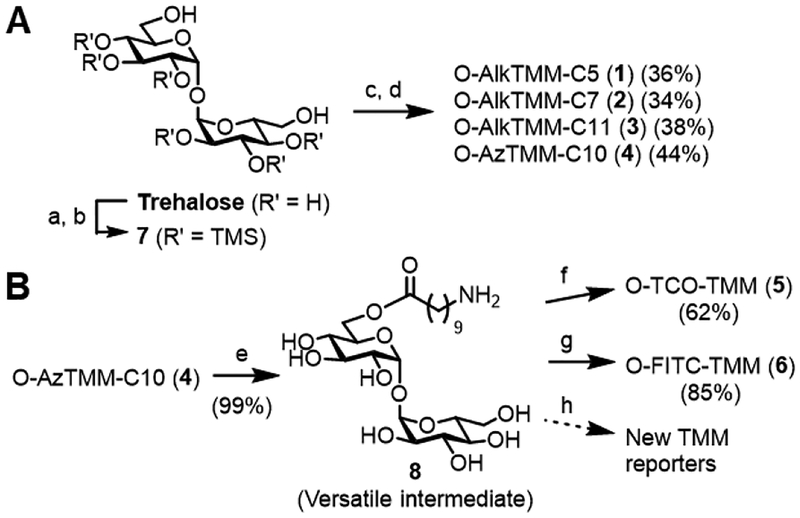
The objective of this study was to develop and characterize an expanded toolbox of TMM-based reporters that are designed to provide broad experimental flexibility for studying the mycomembrane (Figure 1B). First, we designed O-AlkTMM reporters with varying chain lengths, from C5–C11 (1–3), to determine whether this property had any effect on metabolic incorporation efficiency. Second, we designed the azide-modified TMM reporter O-AzTMM-C10 (4) to allow non-toxic Cu-free click chemistry [i.e., strain-promoted azide–alkyne cycloaddition (SPAAC[33,34])] on living cells. O-AzTMM-C10 was also viewed as a versatile synthetic intermediate, since it can be readily elaborated to other TMM analogues via either (i) click chemistry with alkyne-modified reagents or (ii) azide reduction to the corresponding amine, followed by reaction with amine-reactive reagents. Third, we designed the trans-cyclooctene-modified TMM reporter O-TCO-TMM (5) to allow secondary labeling using the tetrazine ligation, which is exceptionally fast and can be performed on live cells.[35,36] Fourth, because reporters 1–5 all require two-step labeling, we designed the fluorescein-modified TMM reporter O-FITC-TMM (6) to allow one-step labeling of live cells. The diversity of TMM modifications explored in this work also allowed for an in-depth evaluation of the tolerance of mycoloylation machinery for varying size and polarity of the tag.
To synthesize compounds 1–4, we used Kulkarni’s approach to accessing unsymmetrical trehalose esters (Scheme 1A).[37] First, trehalose was subjected to per-O-trimethylsilylation, then both 6-O-trimethylsilyl groups were selectively removed using K2CO3/CH3OH to give diol 7. This C2-symmetric intermediate was desymmetrized in the subsequent step, which entailed 6-O-monoesterification with an appropriate alkyne- or azide-modified carboxylic acid in the presence of N,N’-dicyclohexylcarbodiimide (DCC) and catalytic 4-(dimethylamino)pyridine (DMAP). The intermediate was then desilylated in the presence of Dowex H+ resin, delivering the desired products. The yields for compounds 1–4 over the two-step monoesterification–deprotection sequence ranged from 34–44%, consistent with prior results from Kulkarni[37] and our lab.[26]
Scheme 1.
(A) Syntheses of alkyne- and azide-modified TMM reporters 1–4. a) TMSCl, Et3N; b) K2CO3, CH3OH (92% over two steps, ref. 36); c) carboxylic acid, DCC, DMAP, CH2Cl2; d) Dowex H+ resin, CH3OH (yields over two steps are given). (B) Syntheses of TCO- and fluorescein-modified TMM reporters 5 and 6. e) Pd/C, H2, CH3OH; f) NHS-TCO, Et3N, N,N’-dimethylformamide (DMF); g) FITC, Et3N, DMF; h) reaction of 8 with commercially available amine-reactive reagents will provide access to custom TMM reporters.
To access trans-cyclooctene- and fluorescein-modified TMM analogues 5 and 6, we sought to capitalize on the commercial availability of amine-reactive versions of these tags (Scheme 1B). Therefore, we converted O-AzTMM-C10 (4) into the corresponding amine 8 by Pd-catalyzed hydrogenation, which proceeded in nearly quantitative yield (99%). Next, we reacted amine 8 with either the N-hydroxysuccinimidyl carbonate of trans-cyclooctene (NHS-TCO) to generate O-TCO-TMM (5) or fluorescein isothiocyanate (FITC) to generate O-FITC-TMM (6). Purification by C18 reverse-phase chromatography yielded 5 and 6 in 62% and 85% yield, respectively. Conveniently, intermediate 8 allows modification of the TMM scaffold with virtually any type of amine-reactive chemical cargo, which in the future will provide ready access to TMM reporters tailored to specific applications.
Effect of Acyl Chain Length on Incorporation Efficiency
With compounds 1–6 in hand, our first objective was to evaluate the O-AlkTMM series (1–3) to determine whether changes in chain length had an impact on labeling efficiency. Native mycolic acids are α-branched and β-hydroxylated, and they contain anywhere from 22–100 total carbons, depending on the species.[7] The shortened linear chain of our original TMM reporter, O-AlkTMM-C7, was a fairly drastic simplification of the native mycolate, but the compound was still efficiently incorporated into the mycomembrane.
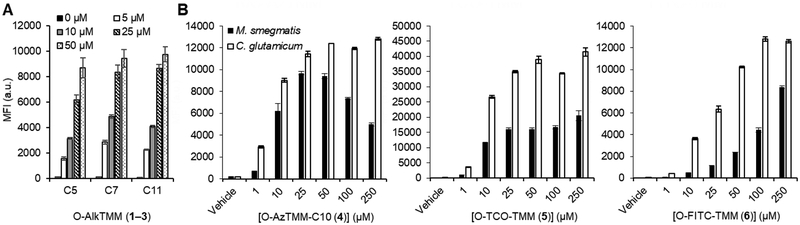
In the present comparison, we predicted to observe that longer-chain reporters would have favorable incorporation. To test this hypothesis, Msmeg was cultured in 0–50 μM of each reporter, then subjected to CuAAC reaction with an azido-488 fluorophore and analyzed by flow cytometry (Figure 2A). All three compounds labeled Msmeg efficiently, which is consistent with the remarkable substrate plasticity that has been reported for mycoloyltransferases.[19,26] Interestingly, at lower probe concentrations (5 and 10 μM), we observed a clear trend in Msmeg in which the intermediate-length O-AlkTMM-C7 (2) gave optimal labeling. One explanation for this observation is that the C7 reporter has an optimal balance of penetration across the mycomembrane and processing by mycoloyltransferases. It is also possible that the smaller C7 reporter labels more terminal AG residues than larger reporters due to a steric advantage.
Figure 2.
(A) Dependence of metabolic labeling on TMM reporter acyl chain length. Msmeg was incubated with 0–50 μM of 1, 2, or 3 (or left untreated) for 4 h, fixed with para-formaldehyde, reacted with Az488 by CuAAC, and analyzed by flow cytometry. (B) Concentration-dependent metabolic labeling of Msmeg and Cglut using azide-, trans-cyclooctene, and fluorophore-modified TMM reporters 4, 5, and 6. Msmeg or Cglut was incubated with 4, 5, or 6 (or left untreated) for 4 h, then in a second step the live cells were reacted with DBCO-488 (for 4), tetrazine-Cy3 (for 5), or left untreated (for 6), then fixed and analyzed by flow cytometry. Error bars denote the standard deviation of three replicate experiments. Y axis on each plot is mean fluorescence intensity (MFI) measured in arbitrary units (a.u.).
It will be interesting in the future to compare reporters 1–3 to versions that more closely resemble the native TMM. For example, compounds with and without α-branching and β-hydroxylation, spanning a broader range of chain length, and containing other mycolic acid modifications would be beneficial to evaluate. To permit such a comparison, strategies for the synthesis of native mycolic acids (e.g., ref. [38] and references therein) will need to be adapted to accomodate alkyne or azide modifications. These studies are ongoing in our lab. In the meantime, there are criteria to be considered for selection of TMM reporter chain length for a given application. The smaller C5 and C7 reporters are conveniently water-soluble and possibly have better porin-mediated uptake across the mycomembrane, whereas larger C11 reporters need co-solvent (e.g., DMSO) to solubilize and may not cross the mycomembrane as efficiently. Metabolic incorporation of the C7 reporter was also significantly higher than either the C5 or C11 compounds. On the other hand, the TMM reporters with longer chains that more closely resemble native mycolates are also more likely to faithfully replicate the biophysical properties of native TMM (and labeled TDM, AGM, or O-mycoloylated protein products) within a cellular environment. These considerations could prove particularly relevant in the development of mycobacteria-specific detection probes.
Labeling of Live Bacteria with TMM Reporters
Due to the complications of using copper in vivo, alkyne-labeled cells (e.g., O-AlkTMM-labeled bacteria) are typically chemically fixed prior to carrying out CuAAC reactions on the cell surface. Fixation prevents studies in live cells, whether in broth culture or more complex settings such as macrophage or animal infection models. To address this issue, the new TMM reporters 4–6 were designed to enable use in living cells, which we investigated next. Each reporter was evaluated over the same concentration range (0–250 μM) in Msmeg and Cglut (Figure 2B). O-AzTMM-C10 (4) labeling was followed by SPAAC reaction on live cells with a dibenzocycloocytne (DBCO)-488 fluorophore; O-TCO-TMM (5) labeling was followed by tetrazine ligation on live cells with a tetrazine-Cy3 fluorophore; since O-FITC-TMM (6) directly delivers the fluorophore during incorporation, no secondary step was required for this reporter. Cellular fluorescence was quantified by flow cytometry. Consistent with prior results from O-AlkTMM labeling experiments, all three reporters efficiently labeled both Msmeg and Cglut as compared to the control cells. Labeling saturation was observed for both O- AzTMM-C10 (4) and O-TCO-TMM (5) at approximately 25 μM concentration, whereas for O-FITC-TMM saturation occurred at higher concentrations (≥ 100 μM), presumably because the larger size of the fluorophore impeded access to mycoloyltransferases or curbed incorporation by mycoloyltransferases. Interestingly, the reporters with the larger TCO and FITC tags, 5 and 6, labeled Cglut much more efficiently than Msmeg. This could be due to increased uptake of the larger reporters across the mycomembrane of Cglut, which has shorter mycolic acids and higher fluidity than Msmeg.[21]
Notably, we did not observe growth inhibition or reduced viability upon treatment of Msmeg with compounds 1–6 at concentrations up to 100 μM over 16 h, demonstrating their apparent non-perturbing nature under these conditions (Figure S1). Interestingly, at 250 μM concentration, partial growth inhibition was observed for O-AlkTMM-C11 (3), O-AzTMM-C10 (4), and O-TCO-TMM (5), all of which have longer acyl chains. The azido TMM analogue had the most significant impairment of Msmeg growth, which likely explains why this compound showed reduced labeling of Msmeg at higher concentrations (Figure 2B). Further investigation of the inhibitory properties of TMM analogues is a topic of future interest.
Thus, in contrast to the original reporter O-AlkTMM-C7, the new TMM reporters 4–6 provide the ability to probe incorporation processes in live bacteria. Of course, O-FITC-TMM (6), or related fluorophore-modified TMM reporters (which can be readily accessed from 8 as shown in Scheme 1B), should be used when a one-step live-cell labeling workflow is desired. For example, one-step labeling may be preferred for its simplicity (e.g., fewer reagents and washes) or when there is concern about whether the secondary fluorophore has sufficient access to the tagged biomolecule. Two-step labeling can be desirable for many reasons, including generally higher metabolic incorporation efficiency and the versatility to deliver virtually any type of secondary reagent to the tagged biomolecule without having to synthesize and evaluate a new TMM analogue.[11]
Comparison of SPAAC and Tetrazine Ligation
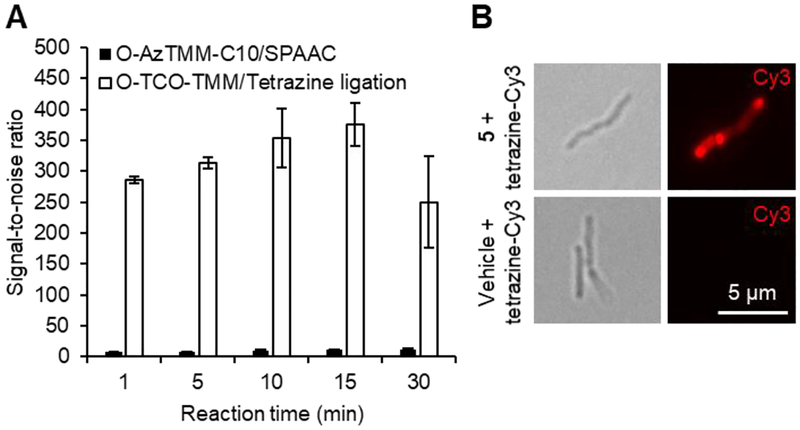
The TMM reporters that allow two-step live-cell labeling workflows, O-AzTMM-C10 (4) and O-TCO-TMM (5), are both useful in these scenarios, although we were interested in characterizing these reporters further to guide probe selection in future applications. TCO–tetrazine ligations, with rate constants ranging from 103–106 M–1s–1, are the fastest bioorthogonal reactions, making them 3–6 orders of magnitude faster than SPAAC reactions employing DBCO, which have rate constants of approximately 0.3 M–1s–1.[39] Thus, we predicted that O-TCO-TMM (5), in combination with the tetrazine ligation, would give optimal cell-surface labeling.
To compare the efficiency of cell-surface labeling using SPAAC or the tetrazine ligation, Msmeg was treated with either 4 or 5 and then reacted with DBCO-Cy3 or tetrazine-Cy3, respectively, for reaction times ranging from 1–30 min. The signal-to-noise (S/N) ratios for each condition, which were determined by flow cytometry, are shown in Figure 3A (signal-to-noise values provide a clearer comparison than mean fluorescence intensities). O-TCO-TMM-labeled cells could be detected by tetrazine ligation with a S/N ratio of nearly 300:1 at only 1 min (Figures 3A and 3B). O-AzTMM-C10-labeled cells could also detected in 1 min by SPAAC, although the S/N ratio was much lower at approximately 10:1. Overall, O-TCO-TMM, in conjunction with the tetrazine ligation, is considered to be an excellent reporter for rapid two-step labeling in living systems, and it should prove useful in situations where very low concentrations of reagents are needed (e.g., in animal infection models). Also, O-TCO-TMM did not exhibit decreased labeling when used at higher concentrations, whereas O-AzTMM-C10 did (Figure 2B). However, O-TCO-TMM does have minor trade-offs, including: (i) the TCO group can isomerize to the unreactive cis isomer over time; and (ii) at present, the availability of tetrazine reagents is still somewhat limited compared to cyclooctyne reagents.
Figure 3.
(A) Cell-surface reaction kinetics comparison between O-AzTMM-C10 (4) with SPAAC and O-TCO-TMM (5) with tetrazine ligation. Msmeg was cultured in 4 or 5 (25 μM each) or left untreated, then reacted with DBCO-Cy3 or tetrazine-Cy3 (20 μM each) for varying times and analyzed by flow cytometry. Signal-to-noise ratios were determined by dividing the MFI value for the reporter-treated sample by the MFI value for the vehicle-treated control exposed to the same bioorthogonal reaction conditions. Error bars denote the standard deviation of three replicate experiments. (B) Fluorescence imaging of O-TCO-TMM-labeled Msmeg after 1 min tetrazine reaction. Scale bar (5 μm) applies to all images. Left, transmitted light images; right, fluorescence images.
Specificity of TMM Reporters for the Mycomembrane
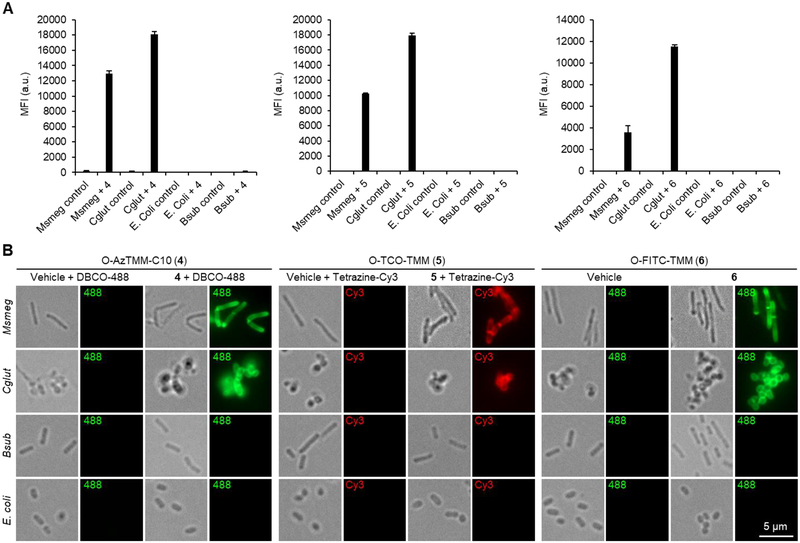
Only species in the Corynebacterineae suborder possess the mycomembrane and its associated biosynthetic pathways. We previously showed that O-AlkTMM-C7 (2) efficiently labeled Msmeg and Cglut but not canonical Gram-positive or Gram-negative bacteria, thus demonstrating that TMM-based reporters are specific for members of the Corynebacterineae suborder.[26] To assess the specificity of the new TMM reporters 4–6 for mycomembrane-containing bacteria, we performed labeling experiments essentially as described above in Msmeg, Cglut, Bacillus subtilis (Bsub, a model Gram-positive organism), and Escherichia coli (E. coli, a model Gram-negative organism). Cellular fluorescence was analyzed by flow cytometry and fluorescence microscopy (Figure 4). These data indicated that the three new TMM reporters exhibited high specificity for members of the Corynebacterineae, with robust labeling of Msmeg and Cglut but no labeling of Bsub or E. coli (Figure 4A). In addition, the microscopy images in Figure 4B showed that treatment with compounds 4–6 resulted in cell-surface fluorescence concentrated at the poles and septa of bacteria, which is consistent with a metabolic route of incorporation, the polar growth mode of Corynebacterineae, and with prior imaging studies employing O-AlkTMM-C7 (2).[23][40]
Figure 4.
Specificity of TMM reporters 4–6 for mycomembrane-containing members of the Corynebacterineae suborder. Msmeg, Cglut, Bsub, or E. coli were incubated with 4 (25 μM), 5 (25 μM), 6 (100 μM), or left untreated for 4 h, then the cells were reacted with DBCO-488 (for 4), tetrazine-Cy3 (for 5), or left untreated (for 6), and analyzed by (A) flow cytometry and (B) fluorescence microscopy. In (A), error bars denote the standard deviation of three replicate experiments. In (B), scale bar (5 μm) at lower right applies to all images; grayscale images are transmitted light images.
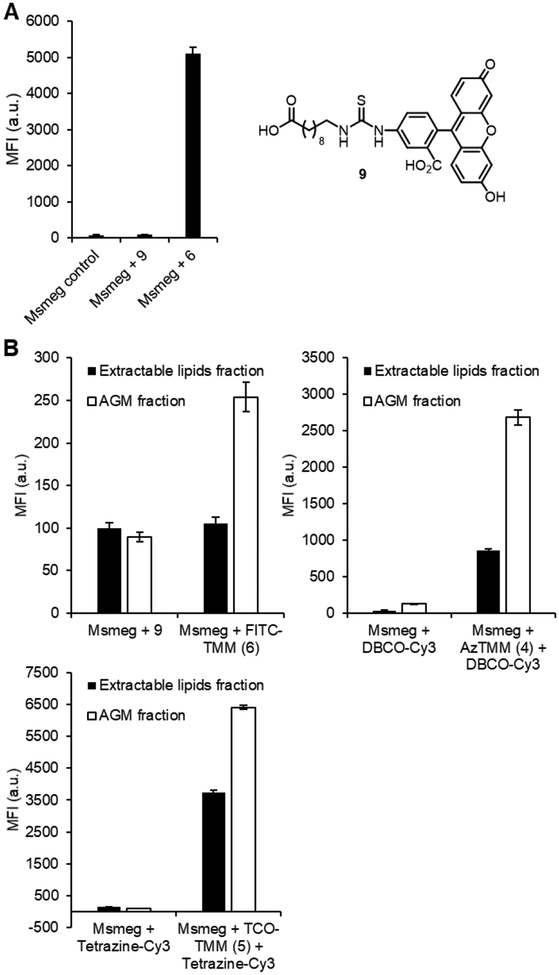
Out of the new TMM reporters 4–6, O-FITC-TMM (6) represents the most significant structural departure from the original reporter O-AlkTMM-C7 (2). We considered the possibility that the fluorescein group could promote non-specific association of 6 with the cell surface rather than metabolic incorporation. To test whether the trehalose ester moiety of O-FITC-TMM was in fact responsible for targeting it to the cell as designed, we also synthesized its corresponding FITC-modified carboxylic acid lacking the ester-linked trehalose (compound 9; see SI for synthesis) (Figure 5A). In a head-to-head comparison, O-FITC-TMM labeled Msmeg efficiently, while compound 9 gave no labeling above the untreated control (Figure 5A), supporting the proposition that O-FITC-TMM incorporation is dependent on trehalose metabolism.
Figure 5.
(A) Cellular labeling comparison of O-FITC-TMM (6) and its trehalose-deficient variant 9. Msmeg was incubated with 6 or 9 (100 μM), or left untreated, then analyzed by flow cytometry. Error bars denote the standard deviation of three replicate experiments. (B) Labeling distribution of compounds 4–6 in different cellular fractions. Msmeg was incubated with 4 (25 μM), 5 (25 μM), 6 (100 μM), 9 (100 μM), or left untreated for 4 h, then reacted with an appropriate fluorophore (if necessary), fractionated into an AGM-containing fraction and extractable lipids, and analyzed using a fluorescence plate reader. Error bars denote the standard deviation of three replicate experiments.
As noted above and depicted in Figure 1A, TMM reporters have the potential to label both major mycomembrane components, including inner-leaflet AGM and outer-leaflet TDM.[41] We previously determined that O-AlkTMM-C7 mainly incorporated into the AGM-containing cellular fraction from Msmeg and less so into the TDM-containing extractable lipids fraction, which is consistent with AGM being the most abundant mycolate-containing envelope component.[26,30] Here, we performed the same cellular fractionation experiments following treatment of Msmeg with 4–6. In agreement with the O-AlkTMM-C7 results, all three new reporters showed the majority of fluorescence localized to the AGM-containing fraction (Figure 5B). It was interesting to note that O-FITC-TMM (6) appeared to give a smaller increase in fluorescence versus the control compound (9) in this fractionation experiment as compared to the cellular labeling experiment, although the reason for this is unclear (Figure 5A).
Next, we sought to test whether incorporation of the new TMM reporters 4–6 into mycomembrane-containing bacteria was dependent on mycoloyltransferase activity. This is a challenging task because all members of the Corynebacterineae have multiple mycoloyltransferase isoforms, which are collectively essential for viability and exhibit high functional redundancy.[15] Not only does this make the knockout of all mycoloyltransferases impossible due to lethality, but experiments involving partial blockage of mycoloyltransferase activity (e.g, by chemical inhibition or partial knockout) are complicated due to isoform redundancy and the resulting perturbation of cell envelope structure and permeability,[42] which presumably can impact probe uptake and thus labeling efficiency. We attempted to use the Ag85C inhibitor ebselen[26,43] and previously reported Msmeg mutants partially lacking mycoloyltransferase activity (e.g., Msmeg ag85a::Tn[44] and the triple knockout Msmeg ΔMSMEG_6396–99[25]), but the results were inconclusive. For example, we observed increases in cellular labeling for all of the TMM reporters in both Msmeg mutants, perhaps suggesting that partial mycoloyltransferase disruption leads to increased reporter uptake, followed by envelope incorporation via the remaining mycoloyltransferases.
Given the complexities posed by the chemical inhibition and Msmeg mutant experiments, we conceived of an alternative approach to determine whether cellular incorporation of TMM-based reporters is mycoloyltransferase-dependent. Cglut encodes six mycoloyltransferases, only one of which, CgMytC, transfers mycoloyl groups from TMM onto polypeptides to generate O-mycoloylated proteins (see Figure 1A).[45] Our group previously showed that O-AlkTMM-C7 could be used to metabolically label O-mycoloylated proteins in Cglut with alkyne tags, enabling their subsequent visualization using CuAAC followed by SDS-PAGE with in-gel fluorescence analysis.[30] We reasoned that using this experimental platform with a Cglut mutant lacking CgMytC would allow us to assess whether TMM reporters operate in a mycoloyltransferase-dependent manner, because (i) CgMytC is not essential; and (ii) this system eliminates the issue of mycoloyltransferase redundancy—again, available evidence suggests that only CgMytC adds O-mycolates to proteins in Cglut. Thus, we expected it would provide a facile readout for specific probe incorporation.
To test this CgMytC deletion approach in Cglut, we regenerated the CgMytC-deficient mutant Cglut Δcg0413 reported in the literature[45] by homologous recombination and confirmed the absence of the gene by both PCR and sequencing (Table S1 and Figure S2). Cglut wild type and the Δcg0413 mutant were incubated in the presence of O-AlkTMM-C7 (2) and the new TMM reporters 4–6. After washing the cells, aliquots for each condition were collected, secondarily labeled with a fluorophore using the appropriate conditions (if necessary), and analyzed by flow cytometry. As depicted in Figure 6A, cellular fluorescence analysis was consistent with our aforementioned Msmeg mutant results—the Δcg0413 mutant showed increased fluorescence for all of the probes compared to wild-type Cglut. As suggested above for the Msmeg mutants, we speculate that this increased labeling in the CgMytC mutant was due to a combination of increased cellular permeability and the activity of the remaining mycoloyltransferases in the mutant. Next, we lysed the remaining TMM reporter-treated cells to collect proteins. The lysates were subjected to appropriate secondary fluorophore labeling (if necessary), then resolved by SDS-PAGE and analyzed by in-gel fluorescence (Figure 6B). O-AlkTMM-C7 and O-AzTMM-C10 showed robust 488 fluorescence for numerous proteins in the 10–75 kDa range, consistent with our prior work with O-AlkTMM-C7,[30] and labeling was completely abolished in the Δcg0413 mutant. The similar probe O-AlkTMM-C11 gave results identical to those obtained for O-AlkTMM-C7 and O-AzTMM-C10 (data not shown). Likewise, the tetrazine ligation-capable reporter O-TCO-TMM exhibited CgMytC-dependent protein labeling, with a major band in the 10–15 kDa region (similar to the O-AlkTMM and O-AzTMM compounds) and lower fluorescence associated with larger proteins, perhaps because this larger probe selectively labels smaller proteins. These data suggest that the O-AlkTMM series, O-AzTMM-C10, and O-TCO-TMM specifically incorporate into mycolate-containing envelope components via mycoloyltransferase activity, as hypothesized.
Figure 6.
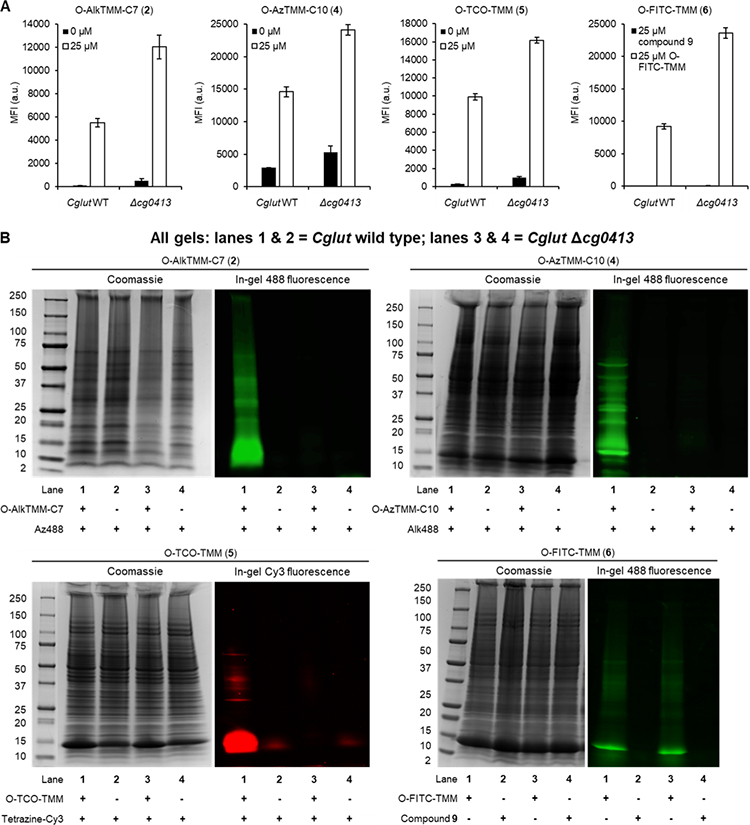
Mycoloyltransferase dependence of labeling Cglut cells (A) and Cglut O-mycoloylated proteins (B) with TMM reporters. Cglut wild type or Δcg0413 mutant were incubated overnight with 100 μM 2, 4, 5, 6 (or 9), or left untreated, then aliquots of cells were removed and reacted with Az488 (for 2), Alk488 (for 4), tetrazine-Cy3 (for 5), or left untreated (for 6 and its corresponding control compound 9), and analyzed by flow cytometry. Error bars denote the standard deviation of three replicate experiments. From the remaining cells, lysates were collected and reacted with the same secondary reagents (if needed), then analyzed by SDS-PAGE. For each probe evaluated, lanes 1 and 2 are from Cglut wild type and lanes 3 and 4 are from Cglut Δcg0413; the Coomassie-stained gel is shown on the left, and in-gel fluorescence is shown on the right.
O-FITC-TMM also labeled Cglut proteins (Figure 6B), but this signal was present in both wild-type Cglut and the Δcg0413 mutant, implying a lack of CgMytC dependence. Our other data (e.g., Figures 4, 5, and 6A) demonstrate consistent behavior between O-FITC-TMM and the other TMM reporters, so it is possible that O-FITC-TMM is not a good substrate for CgMytC but it is nonetheless efficiently processed by other glycolipid-synthesizing mycoloyltransferases. Regardless, based on the Cglut protein labeling results (Figure 6B), it cannot be ruled out that O-FITC-TMM incorporates into mycomembrane-containing bacteria by a mycoloyltransferase-independent route. Taken together, the new TMM reporters described in this study generally behaved like the previously reported O-AlkTMM-C7 compound with respect to their mycomembrane specificity. However, because we could not confirm that O-FITC-TMM labeled bacteria via mycoloyltransferase activity, at present this compound should be limited to applications where mycobacteria-specific surface labeling is desired but the labeling route is inconsequential. It will be of interest to further explore the ramifications of adding large fluorophores (or other sizeable chemical cargo) to the TMM scaffold in follow-on studies. For instance, it was recently shown that D-amino acid-based reporters of peptidoglycan synthesis incorporate via different labeling routes depending on the nature of the detectable tag.[40]
Conclusions
In summary, we reported herein the efficient chemical synthesis and evaluation of a collection of TMM-based metabolic reporters that will facilitate research on the mycomembrane. The chemical tags appended to the TMM analogues’ acyl chains allow labeling of mycomembrane-containing bacteria through two-step strategies employing the major bioorthogonal reactions (CuAAC, SPAAC, or tetrazine ligation) or one-step strategies employing TMM reporters bearing pre-attached fluorophores. In addition to providing improved flexibility in experimental design versus the original O-AlkTMM-C7 reporter, the new reporters can be deployed in living mycobacteria. Of particular interest, this is the first method reported for modifying the mycobacterial cell surface using the rapid and highly efficient TCO–tetrazine ligation.
In addition, this study further exemplified the high promiscuity of cellular mycoloylation machinery for unnatural TMM analogues. This feature of mycobacteria, along with the versatile synthetic intermediates (i.e., 4 and 8) and high specificity of TMM-based reporters for mycobacteria reported herein, shows that TMM analogues can be used to develop novel strategies for targeting mycobacteria with various types of chemical payload. Indeed, the TMM scaffold offers opportunities for the development of new mycomembrane-targeting probes. For instance, the Kiessling group recently expanded on the TMM reporter concept with a FRET-based TMM analogue, called QTF, which fluoresced upon Ag85-mediated separation of its trehalose-linked quencher and “lipid“-linked fluorophore.[46] QTF does not label mycomembrane components, rather it allows direct visualization of mycoloyltransferase activity, a novel and attractive feature that complements the TMM reporters developed by our group so far. Such fluorogenic probes that specifically detect mycobacteria in a single step are of interest as tuberculosis diagnostic tools,[25] and trehalose ester-based probes warrant further investigation in this regard.
This study also revealed cell labeling characteristics of TMM reporters that should be considered when applying this class of compounds. Most significantly, we encountered challenges with establishing metabolic pathway specificity for the new TMM reporters using partial depletion of mycoloyltransferase activity. This issue probably stems from the impact such conditions have on mycomembrane integrity and permeability, which could alter probe uptake. To circumvent this issue, we developed a new system that allowed us to establish pathway specificity by exploiting the Cglut protein O-mycoloylation pathway. However, this issue may complicate the use of TMM reporters to analyze the impact of inhibitors or genetic manipulation on mycoloyltransferase activity in whole cells (e.g., Figure 6A shows that CgMytC deletion gave an unexpected increase in cellular labeling). Indeed, the signal observed from whole-cell labeling experiments results from a combination of probe incorporation via biosynthetic enzymes—and, in principle, probe removal via degradatory enzymes—as well as accessibility of the probe to those enzymes. Additional research is needed to fully investigate this topic and possibly capitalize on it, for example with the development of membrane permeability probes. In the meantime, whole-cell experiments involving partial mycoloyltransferase inactivation should be designed and interepreted cautiously. Irrespective of this issue, TMM-based reporters have numerous capabilities with respect to mycobacteria research, including visualization of envelope dynamics, characterization of mycomembrane proteins, specific detection of mycobacterial cells, and versatile cell-surface engineering of live mycobacteria.
Experimental Section
The Supporting Information contains supplementary results, experimental procedures, and NMR spectra of synthetic compounds.
Supplementary Material
Acknowledgments
B.M.S. was supported by an NSF CAREER Award (1654408), a Cottrell College Scholar Award from the Research Corporation for Science Advancement (22525), and a Henry Dreyfus Teacher-Scholar Award from The Camille & Henry Dreyfus Foundation (TH-17–034). M.S.S was supported by the National Institutes of Health (NIH DP2 AI138238). We thank J. C. Seeliger for helpful discussions. We thank R. J. Hood for assistance with NMR spectroscopy and A. N. Ilacqua for assistance with flow cytometry.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Dye C, Lancet 2006, 367, 938–940. [DOI] [PubMed] [Google Scholar]
- [2].World Health Organization, Global Tuberculosis Report 2017.
- [3].Woo HM, Park J-B, J. Biotechnol 2014, 180, 43–51. [DOI] [PubMed] [Google Scholar]
- [4].Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H, Proc. Natl. Acad. Sci. U.S.A 2008, 105, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M, J Bacteriol 2008, 190, 5672–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barry CE, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y, Prog. Lipid Res 1998, 37, 143–179. [DOI] [PubMed] [Google Scholar]
- [7].Marrakchi H, Lanéelle M-A, Daffé M, Chem. Biol 2013, 21, 67–85. [DOI] [PubMed] [Google Scholar]
- [8].North EJ, Jackson M, Lee RE, Curr. Pharm. Des 2014, 20, 4357–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Slayden RA, Barry CE, Methods. Mol. Med 2001, 54, 229–245. [DOI] [PubMed] [Google Scholar]
- [10].Rezwan M, Lanéelle MA, Sander P, Daffé M, Microbiol J. Methods 2007, 68, 32–39. [DOI] [PubMed] [Google Scholar]
- [11].Siegrist MS, Swarts BM, Fox DM, Lim SA, Bertozzi CR, FEMS Microbiol. Rev 2015, 39, 184–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kalscheuer R, Koliwer-Brandl H, Microbiol. Spectr 2014, 2, DOI: 10.1128/microbiolspec.MGM2-0002-2013. [DOI] [PubMed] [Google Scholar]
- [13].Gavalda S, Bardou F, Laval F, Bon C, Malaga W, Chalut C, Guilhot C, Mourey L, Daffé M, Quémard A, Chem. Biol 2014, 21, 1660–1669. [DOI] [PubMed] [Google Scholar]
- [14].Grzegorzewicz AE, Pham H, Gundi VAKB, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SEM, Kordulakova J, et al. , Nat. Chem. Biol 2012, 8, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dautin N, de Sousa-d’Auria C, Constantinesco-Becker F, Labarre C, Oberto J, de la Sierra-Gallay IL, Dietrich C, Issa H, Houssin C, Bayan N, Biochim. Biophys. Acta - Gen. Subj 2016, 1861, 3581–3592. [DOI] [PubMed] [Google Scholar]
- [16].Sathyamoorthy N, Takayama K, J. Biol. Chem 1987, 262, 13417–13423. [PubMed] [Google Scholar]
- [17].Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS, Science 1997, 276, 1420–1422. [DOI] [PubMed] [Google Scholar]
- [18].Huc E, Meniche X, Benz R, Bayan N, Ghazi A, Tropis M, Daffé M, J. Biol. Chem 2010, 285, 21908–21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D’Hooge F, Lee SS, Via LE, Tahlan K, Barry CE 3rd, et al. , Nat. Chem. Biol 2011, 7, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Swarts BM, Holsclaw CM, Jewett JC, Alber M, Fox DM, Siegrist MS, Leary JA, Kalscheuer R, Bertozzi CR, J. Am. Chem. Soc 2012, 134, 16123–16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rodriguez-Rivera FP, Zhou X, Theriot JA, Bertozzi CR, J. Am. Chem. Soc 2017, 139, 3488–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu Z, Meshcheryakov VA, Poce G, Chng S-S, Proc. Natl. Acad. Sci. U. S. A 2017, 114, 7993–7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rundell SR, Wagar ZL, Meints LM, Olson CD, O’Neill MK, Piligian BF, Poston AW, Hood RJ, Woodruff PJ, Swarts BM, Org. Biomol. Chem 2016, 14, 8598–8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peña-Zalbidea S, Huang AY-T, Kavunja HW, Salinas B, Desco M, Drake C, Woodruff PJ, Vaquero JJ, Swarts BM, Carbohydr. Res 2018, DOI: 10.1016/j.carres.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kamariza M, Shieh P, Ealand CS, Peters JS, Chu B, Rodriguez-Rivera FP, Babu Sait MR, Treuren VW, Martinson N, Kalscheuer R, et al. , Sci. Transl. Med 2018, 10, eaam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Foley HN, Stewart JA, Kavunja HW, Rundell SR, Swarts BM, Angew. Chem. Int. Ed 2016, 55, 2053–2057. [DOI] [PubMed] [Google Scholar]
- [27].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB, Angew. Chem. Int. Ed 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- [28].Tornøe CW, Christensen C, Meldal M, J. Org. Chem 2002, 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- [29].Touchette MH, Van Vlack ER, Bai L, Kim J, Cognetta AB 3rd, Previti ML, Backus KM, Martin DW, Cravatt BF, Seeliger JC, ACS Infect. Dis 2017, 3, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kavunja HW, Piligian BF, Fiolek TJ, Foley HN, Nathan TO, Swarts BM, Chem. Commun 2016, 52, 13795–13798. [DOI] [PubMed] [Google Scholar]
- [31].Issa H, Huc-Claustre E, Reddad T, Bonadé Bottino N, Tropis M, Houssin C, Daffé M, Bayan N, Dautin N, PLoS One 2017, 12, e0171955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rangan KJ, Yang Y-Y, Charron G, Hang HC, J. Am. Chem. Soc 2010, 132, 10628–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Agard NJ, Prescher JA, Bertozzi CR, J. Am. Chem. Soc 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- [34].Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang VP, Miller IA, Lo A, Codelli JA, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Blackman ML, Royzen M, Fox JM, J. Am. Chem. Soc 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Devaraj NK, Weissleder R, Hilderbrand a S., Bioconjugate Chem 2008, 19, 2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sarpe VA, Kulkarni SS, J. Org. Chem 2011, 76, 6866–6870. [DOI] [PubMed] [Google Scholar]
- [38].van der Peet PL, Gunawan C, Torigoe S, Yamasaki S, Williams SJ, Chem. Commun 2015, 51, 5100–5103. [DOI] [PubMed] [Google Scholar]
- [39].Patterson DM, Nazarova LA, Prescher JA, ACS Chem. Biol 2014, 9, 592–605. [DOI] [PubMed] [Google Scholar]
- [40].García-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Chuin Lim H, Wagner JC, Morita YS, Swarts BM, et al. , Elife 2018, 7, e37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chiaradia L, Lefebvre C, Parra J, Marcoux J, Burlet-Schiltz O, Etienne G, Tropis M, Daffé M, Sci. Rep 2017, 7, 12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jackson M, Raynaud C, Lanéelle M-A, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffé M, Mol. Microbiol 1999, 31, 1573–1587. [DOI] [PubMed] [Google Scholar]
- [43].Favrot L, Grzegorzewicz AE, Lajiness DH, Marvin RK, Boucau J, Isailovic D, Jackson M, Ronning DR, Nat. Commun 2013, 4, 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nguyen L, Chinnapapagari S, Thompson CJ, J. Bacteriol 2005, 187, 6603–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huc E, de Sousa-D’Auria C, de la Sierra-Gallay IL, Salmeron C, van Tilbeurgh H, Bayan N, Houssin C, Daffé M, Tropis M, J. Bacteriol 2013, 195, 4121–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hodges HL, Brown RA, Crooks JA, Weibel DB, Kiessling LL, Proc. Natl. Acad. Sci. U. S. A 2018, 115, 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.