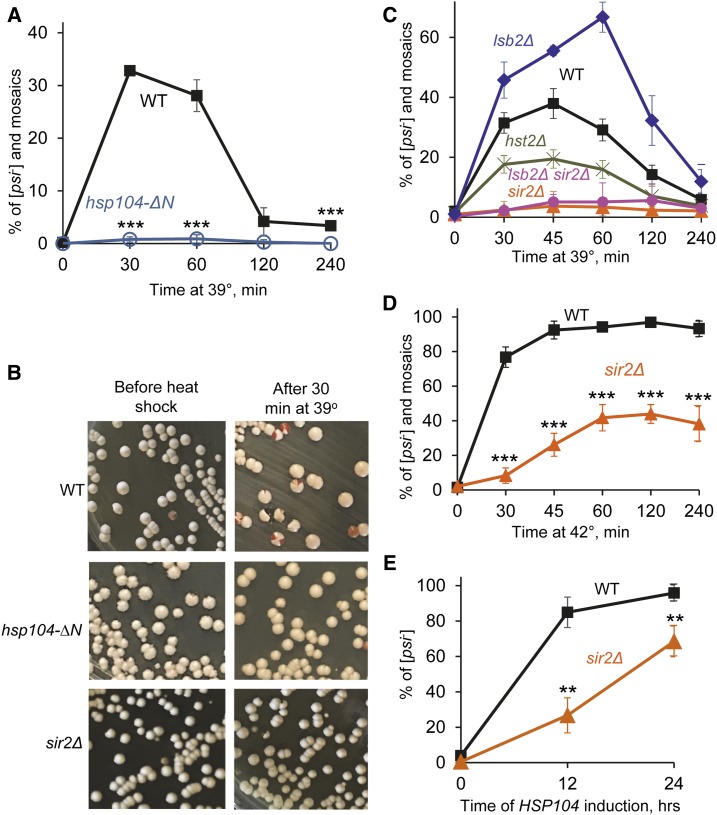
Figure 1.
Effects of the deletions of HSP104N or SIR2 on [PSI+] destabilization. (A) Deletion of HSP104N (hsp104-∆N) prevents [PSI+] destabilization by short-term mild heat shock at 39°, compared to the isogenic wild-type (WT) strain. Numbers of colonies are shown in Table S2. (B) Examples of plates showing [PSI+] destabilization by mild heat shock in the WT strain (as detected by the appearance of red and mosaic pink-red colonies) and lack of destabilization in the hsp104-∆N and sir2∆ strains. (C) Quantitation of [PSI+] destabilization after mild heat shock at 39° in the WT, sir2∆, hst2∆, lsb2∆, and double lsb2∆ sir2∆ strains. Numbers of colonies are shown in Table S3. (D) Deletion of SIR2 (sir2∆) decreases [PSI+] destabilization by heat shock at 42°, compared to isogenic WT strain. Numbers of colonies are shown in Table S5. (E) Sir2∆ delays [PSI+] curing by Hsp104 overexpression, compared to isogenic WT strain. The PGAL-HSP104 construct was induced in galactose medium. Cells were sampled after indicated periods of time and plated onto the synthetic medium selected for the plasmids. The [psi−] colonies were identified after velveteen replica plating to the medium lacking adenine. Numbers of colonies are shown in Table S6. Error bars on A and C–E indicate SD. In general, the differences were statistically significant in all cases where deviations do not overlap. The P values are indicated by asterisks in parts A, D, and E: * P < 0.05, ** P < 0.01, *** P < 0.001. The P values for part C are shown in Table S4.