Abstract
Hybrid male progeny from interspecies crosses are more prone to sterility or inviability than hybrid female progeny, and the male sterility and inviability often demonstrate parent-of-origin asymmetry. However, the underlying genetic mechanism of asymmetric sterility or inviability remains elusive. We previously established a genome-wide hybrid incompatibility (HI) landscape between Caenorhabditis briggsae and C. nigoni by phenotyping a large collection of C. nigoni strains each carrying a C. briggsae introgression. In this study, we systematically dissect the genetic mechanism of asymmetric sterility and inviability in both hybrid male and female progeny between the two species. Specifically, we performed reciprocal crosses between C. briggsae and different C. nigoni strains that each carry a GFP-labeled C. briggsae genomic fragment referred to as introgression, and scored the HI phenotypes in the F1 progeny. The aggregated introgressions cover 94.6% of the C. briggsae genome, including 100% of the X chromosome. Surprisingly, we observed that two C. briggsae X fragments that produce C. nigoni male sterility as an introgression rescued hybrid F1 sterility in males fathered by C. briggsae. Subsequent backcrossing analyses indicated that a specific interaction between the X-linked interaction and one autosome introgression is required to rescue the hybrid male sterility. In addition, we identified another two C. briggsae genomic intervals on chromosomes II and IV that can rescue the inviability, but not the sterility, of hybrid F1 males fathered by C. nigoni, suggesting the involvement of differential epistatic interactions in the asymmetric hybrid male fertility and inviability. Importantly, backcrossing of the rescued sterile males with C. nigoni led to the isolation of a 1.1-Mb genomic interval that specifically interacts with an X-linked introgression, which is essential for hybrid male fertility. We further identified three C. briggsae genomic intervals on chromosome I, II, and III that produced inviability in all F1 progeny, dependent on or independent of the parent-of-origin. Taken together, we identified multiple independent interacting loci that are responsible for asymmetric hybrid male and female sterility, and inviability, which lays a foundation for their molecular characterization.
Keywords: hybrid male sterility, Caenorhabditis briggsae, C. nigoni, introgression, X–autosome interaction
IT is common that closely related species can mate with each other, but their hybrid progeny are often sterile or inviable, especially in the male progeny. The mechanism underlying the asymmetric sterility or inviability remains poorly understood. We previously addressed this question between two nematodes, Caenorhabditis briggsae and C. nigoni, by systematic substitution of various parts of the C. nigoni genome with its C. briggsae’s equivalent followed by phenotypic examination. Here, we investigate the genetic mechanism of the asymmetric sterility and inviability in the hybrid F1 male and female progeny between the two species. We achieved this by crossing a cohort of C. nigoni strains each carrying a substitution with C. briggsae, which led to differential homozygosity of the C. briggsae substitution in the hybrid progeny. The aggregated substitutions covered 94.6% of the C. briggsae genome, including 100% of the X chromosome. Surprisingly, we identified two C. briggsae X fragments that produced C. nigoni male sterility as a substitution but rescued hybrid F1 sterility in males fathered by C. briggsae, indicating that at least two separate X–autosome interactions are involved in the hybrid male sterility. In addition, we identified multiple genomic intervals on C. briggsae autosomes that can rescue the inviability, but not the sterility, of hybrid F1 males fathered by C. nigoni. Importantly, we isolated a 1.1-Mb genomic interval that specifically interacts with an X-linked introgression, which is essential for hybrid male fertility. We further identified three C. briggsae genomic intervals on chromosomes I, II, and III that produce inviability in all F1 progeny, dependent on or independent of the parent-of-origin. The identified interacting loci lay a foundation for their molecular characterization.
Postzygotic hybrid incompatibility (HI) presents one of the major barriers to gene flow between species or populations, leading to reproduction isolation or its enforcement. HI commonly manifests as hybrid male sterility or overall hybrid inviability, and has undergone intensive study in recent decades (Maheshwari and Barbash 2011). The identification of HI loci has become a major task for evolutionary biologists. Antagonistic interactions between parental alleles with differential divergence are predicted to become incompatible, hereafter termed Dobzhansky–Muller incompatibility (DMI) (Muller 1942; Lewontin 1997), which is believed to be responsible for the HI phenotypes in the hybrid progeny. Genetic studies of HI have isolated various loci across phyla. A subset of the HI loci has been molecularly cloned, which has provided unprecedented insights into speciation genetics (Maheshwari and Barbash 2011). Hybrid sterility is more common than other types of HI phenotypes in the heterogametic sex, which is dubbed Haldane’s rule (Schilthuizen et al. 2011). The asymmetry in HI, including male sterility or viability phenotypes, often depends on cross direction, which is called Darwin’s corollary to Haldane’s rule (Turelli and Moyle 2007). The two rules are well supported in the interspecies hybrids cross species (Masly and Presgraves 2007; Woodruff et al. 2010; Kozlowska et al. 2012; Bi et al. 2015). The X chromosome has been shown to play a disproportionately larger role in the hybrid sterility of heterogametic sex in diverse organisms, which is termed the “large-X effect” (Coyne 1985; Presgraves 2008). However, whether X-linked incompatibilities contribute disproportionately to asymmetric hybrid inviability, especially the female’s inviability, has yet been established (Moran et al. 2017). The key to understanding the genetic mechanisms of these two rules is to isolate specific genomic intervals that are required for the asymmetric HI phenotypes in both hybrid F1 and nearly isogenic genetic backgrounds (Coyne and Orr 2004).
C. elegans has been intensively studied for neuron development (Hisamoto and Matsumoto 2017), tissue differentiation (Shao et al. 2013; Gieseler et al. 2017), organogenesis (Mango 2009), and population genetics (Ghosh et al. 2012; Dey et al. 2013; Zamanian et al. 2018). However, it has contributed little to speciation genetics, although incipient speciation seems to be obvious among various populations (Seidel et al. 2008; Ben-David et al. 2017). Such study has been inhibited by the lack of a sister species that can mate with C. elegans and produce viable hybrid progeny (Baird et al. 1992). An improved sampling method introduced in the early 2000s dramatically accelerated the recovery of new Caenorhabditis species (Kiontke et al. 2011; Félix et al. 2014; Huang et al. 2014), which led to the discovery of a few new pairs of sister species that can mate and produce viable progeny (Woodruff et al. 2010; Bundus et al. 2018). For example, crosses between a newly identified sister species pair, C. remanei and C. laten, produced pronounced asymmetric hybrid male sterility and extensive breakdown of backcrossing progeny with hybrid F1 worms (hereafter termed “B2”) (Dey et al. 2014). Reciprocal crosses suggested that the genetic basis of hybrid inviability is more complex than hybrid male sterility and that hybrid male sterility in nematodes involves a single X–autosome interaction (Bundus et al. 2018). Unfortunately, the mapping resolution of HI in hybrid F1 between the two species is relatively coarse.
The identification of the C. briggsae sister species, C. nigoni, has paved the way for the use of this pair for speciation genetics (Woodruff et al. 2010). C. briggsae is a close relative of C. elegans. They share morphology and developmental patterns (Zhao et al. 2008), and are mostly hermaphrodites with occasional males, whereas C. nigoni is a strictly dioecious species that is mostly found in tropical areas (Woodruff et al. 2010). Both species contain five autosomes and a single sex chromosome, X, with XO as male, and XX as female or hermaphrodite. High-quality genomes have been produced for both species (Ross et al. 2011; Ren et al. 2018; Yin et al. 2018). Notably, the genome sizes between the two species differ significantly from each other. For example, the size of the C. briggsae genome is ∼108 Mb with an X chromosome of 21.5 Mb, whereas the size of the C. briggsae genome is ≤ 130 Mb with an X chromosome of 23.6 and 27.3 Mb, depending on DNA-sequencing and genome assembly methodologies. Studies on their hybrids have supported both Haldane’s rule and Darwin’s corollary to Haldane’s rule (Woodruff et al. 2010; Kozlowska et al. 2012). For example, a cross with C. briggsae as the father produces hybrid fertile F1 females but sterile F1 males, whereas a cross in the opposite direction produces hybrid fertile F1females only (Figure 1). However, the genetic mechanism that underlies the asymmetric HI phenotypes remains elusive. To empower C. briggsae and C. nigoni as a model for speciation genetics, we previously generated ∼100 visible transgenic markers that express green fluorescent protein (GFP) and each was inserted into a different part of an individual chromosome (Yan et al. 2012; Bi et al. 2015). Because they can serve as a dominant marker for the linked C. briggsae genomic fragment in the hybrid progeny during backcrossing, these markers have greatly facilitated mapping of the HI loci between the two species. Systematic backcrossing of all of these markers into C. nigoni for ≥ 15 generations has led to a genome-wide HI landscape consisting of C. briggsae introgression in an otherwise C. nigoni background (Bi et al. 2015). Notably, these HI loci were all identified in an essentially C. nigoni background. It is expected that the underlying mechanisms of HI in hybrid F1 worms and in the essentially isogenic genetic backgrounds that carry an introgression will differ greatly, but will also remain poorly defined in any species.
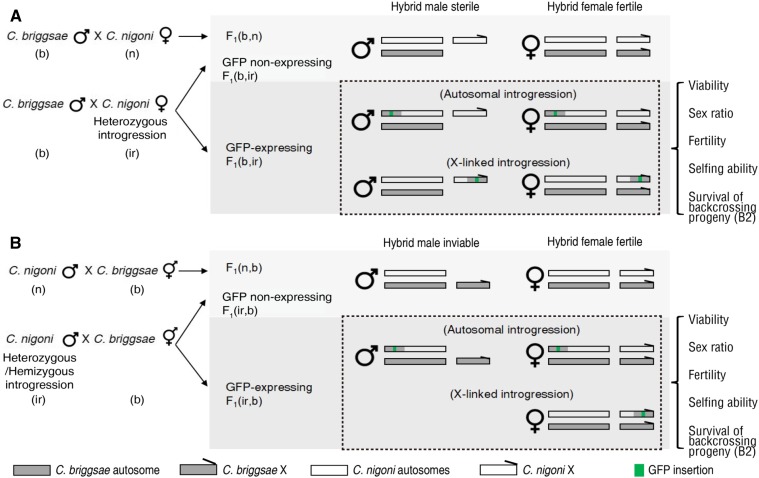
Figure 1.
Strategy for screening for changes in hybrid-incompatible (HI) phenotypes in the F1 progeny from crosses between wild-type C. briggsae and C. nigoni introgression lines, as opposed to the F1 progeny from crosses between wild isolates of two parental species. Shown are schematics of crosses with C. briggsae (b) wild isolate (n) as a father (A) or a mother (B). Crosses with C. nigoni wild isolate (n) or its introgression line (ir) are shown on the top and bottom, respectively. GFP-linked introgression on autosomes or the X chromosome are shown separately. HI phenotypes were scored for the F1 GFP-expressing (introgression-bearing) progeny as indicated. Survival of B2 progeny in the crosses between the hybrid F1 progeny and C. briggsae wild isolate was also counted. Cross progeny were named as described previously (Woodruff et al. 2010). Briefly, progeny were named after their genotypes in parentheses, with paternal and maternal parents listed on the left and right, respectively.
Previous studies have shown little involvement of mitochondria in the hybrid F1 phenotype (Bundus et al. 2015), indicating that most HI phenotypes result from incompatibility between nuclear genomes. A loss-of-function allele in Cbr-him-8 in C. briggsae hermaphrodites seemed to produce viable and fertile hybrid male progeny when mated to the C. nigoni father (Ragavapuram et al. 2016). Such males would allow many other genetic studies, such as the isolation of genes responsible for hermaphroditism. However, a subsequent repeat of this cross experiment was unsuccessful (Ryan and Haag 2017), suggesting that the rescue of the male sterility and inviability may be an artifact from incomplete sperm depletion of the C. briggsae mother. To search for the rescue of male sterility or inviability observed from reciprocal crosses between the two species, we took advantage of a large collection of C. nigoni strains each carrying an C. briggsae-specific introgression fragment that we had generated previously (Bi et al. 2015). We performed a genome-wide screen for specific C. briggsae genomic fragments that could rescue the male sterility or inviability, by crossing the introgression-bearing C. nigoni with C. briggsae in both directions whenever applicable. We identified various introgression fragments that were able to rescue either the male sterility or inviability. We also identified genomic intervals that killed both hybrid males and females in a cross direction-dependent or -independent way. The availability of hybrid F1 fertile males further permitted us to isolate a specific X–autosome interaction that is essential for hybrid male fertility.
Materials and Methods
Nematode strains and maintenance
The following strains were used in the study: C. briggsae (AF16), C. nigoni (JU1421), C. nigoni cytoplasm-replaced hybrids (ZZY10357), and 29 C. nigoni strains carrying a GFP-flagged C. briggsae introgression (Supplemental Material, Table S1). ZZY10357 worms carrying a complete set of C. nigoni chromosomes and C. briggsae cytoplasm were generated by backcrossing C. briggsae hermaphrodites with C. nigoni male worms for 20 generations. Therefore, only mitochondria from C. briggsae but not from C. nigoni are present in the C. nigoni background, as judged by genotyping with C. briggsae-specific PCR primers. All strains were maintained at a constant temperature of 25° on 1.5% agar NGM (nematode growth medium) plates seeded with the Escherichia coli strain OP50.
Cross setups and nomenclature of hybrids
The paternal and maternal strains were named as described previously (Woodruff et al. 2010). Specifically, the genotypes of the hybrid F1 progeny were indicated by their parents-of-origin in parentheses after F1, which were separated by a comma. The letters “b” and “n” were used to denote C. briggsae and C. nigoni, respectively. The word “ir” was short for introgression and a five-digit number derived from its corresponding strain name was used to denote a specific strain. For, example, F1(b, n) represents F1 hybrids from a cross between C. briggsae male and C. nigoni female animals, while F1(n, b) represents F1 hybrids from the opposite cross direction. When introgression-bearing C. nigoni male/female animals were crossed with C. briggsae hermaphrodite/male animals, their hybrids were referred to as F1(ir, b) and F1(b, ir), respectively. For a specific introgression, a unique number derived from its name (Table S1) was used to replace ir. For example, F1(b, 10330) denotes hybrid F1 progeny derived from the cross between a C. briggsae male and C. nigoni female carrying introgression zzyIR10330.
Viability
Hybrid viability was measured as the number of hybrids could that survive to adulthood. Specifically, F1(b, ir) or F1(ir, b) hybrid viability was measured by scoring the number of GFP-expressing, and non-GFP-expressing, male and female adults, which was used to evaluate the effect of an introgression on the hybrid viability relative to that of hybrid F1(b, n) or F1(n, b) worms. Note that hybrid inviability was exclusively found in F1(n, b) male hybrids, but not in F1(b, n) male or female hybrids from both cross directions.
Male ratio
Given that there were some intraspecific variations between male ratios scored using different strains (Kozlowska et al. 2012), male ratios were scored in hybrids derived from crosses between wild isolates of C. briggsae (AF16) and C. nigoni (JU1421) in both cross directions as the control. For an introgression of interest, the hybrid male ratio was scored as the proportion of hybrid male adults expressing GFP out of all GFP-expressing hybrid progeny. The final ratio was calculated from the average of at least three replicates, each being scored for ≥ 150 animals.
Fertility
Hybrid fertility was defined as the ability to produce embryos after mating with the opposite sex from either C. briggsae or C. nigoni. Note that hybrid sterility was exclusively found in F1(b, n) males, but not in F1(b, n) or F1(n, b) females (Figure 1) (Woodruff et al. 2010). F1(n, b) males were unsuitable for fertility tests due to hybrid inviability, except in the case of rescued inviability. The fertility of hybrid males bearing an introgression of interest was evaluated by mating GFP-expressing males with C. nigoni L4 females or C. briggsae sperm-depleted hermaphrodites for 24 hr. The fertility of hybrid females bearing an introgression of interest was identified by mating GFP-expressing L4 females with C. briggsae or C. nigoni males for 24 hr. Fertility was quantified as brood size, which as calculated from the average number of embryos produced by at least three individual worms over their whole life span.
Screen for selfing ability
C. nigoni carrying an introgression (GFP-expressing) were mated with C. briggsae for 12 hr in both directions. Parents were killed. Laid eggs were allowed to develop to the L4 stage followed by the transfer of individual females to separate plates. The numbers of embryos and animals grown were counted after 72-hr cultivation on NGM plates with food. The B2 animals were mated with C. briggsae and C. nigoni for fertility.
Survival rate of backcrossing B2 worms
In a cross between F1 female hybrids and C. briggsae males, hundreds of backcrossing B2 embryos were laid but < 1% of them were able to develop to adulthood. The cross was set up by mating GFP-expressing F1(b, ir) or F1(ir, b) L4 females with C. briggsae males for 24 hr, followed by transfer of the F1 female hybrids every 12 hr until they stopped laying eggs. The numbers of embryos and surviving adults were counted. The survival rate was calculated as the average ratio of B2 adults out of the total B2 embryos in three replicates, with ≥ 150 embryos counted in each replicate.
Microscopy
A Leica LSM 510 confocal laser microscope was used to take all DIC micrographs. Worms were paralyzed in M9 buffer with 0.05 M sodium azide. Spermatids from dissected worms were incubated in sperm medium (SM) containing 50 mM HEPES pH 7.8, 50 mM NaCl, 25 mM KCl, 5 mM CaCl2, 1 mM MgSO4, and 1 mg/ml BSA (Nelson and Ward 1980).
Sperm size quantification
GFP-expressing well-fed males were picked under a stereo microscope. Spermatids were obtained by dissecting male worms in SM followed by imaging with the Leica microscope. ImageJ (Rueden et al. 2017) was used to automatically measure cross-sectional areas of spermatids using the function “particle analysis.” For each experiment, sperm sizes were measured for three or four males, each producing 85–648 spermatids.
Sperm activation and its quantification
Sperm were activated in freshly made SM containing diluted pronase (Sigma [Sigma Chemical], St. Louis, MO) in a final concentration of 200 ng/μl. Micrographs were taken within 5–30 min of dissection. The activation rates of wild-type C. briggsae or C. nigoni male sperm were scored as controls. Sperm activation rates were calculated as a percentage of the average activation rate (number of activated spermatids divided by the number of all sperm in a microscopic view) of three or four male adults.
Nanopore sequencing for genotyping zzyIR10230 that rescued the male sterility of ZZY10330
Fertile GFP-expressing F20 male hybrids were mated with L4 stage C. nigoni female worms in three batches. Genomic DNA was extracted from the females of 21st generation backcrossing progeny. A total of 371 F21 adult females were picked for DNA extraction and Nanopore pore sequencing as described previously (Ren et al. 2018). GFP-expressing F20 male hybrids were genotyped with PCR to ensure that the boundary of X-linked introgression was the same with zzyIR10330, which produced the male sterility in C. nigoni background. It was expected that 100% of the F21 female worms would carry the GFP-flagged zzyIR10330 as a result of sex-linked segregation of the introgression, and that 50% would carry the non-GFP-flagged zzyIR10230 as a heterozygote.
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures and supplemental figures, and tables and supplemental tables. Table S1 lists the details of the introgression strains used in hybrid F1 phenotypic screening. Table S2 lists the newly designed genotyping primers based on C. briggsae genome assembly (cb4) in addition to those listed previously (Bi et al. 2015). Table S3 shows the statistics of hybrid progeny resulting from crosses that produced complete inviability. The Nanopore sequencing data were submitted to the National Center for Biotechnology Information under BioProject number PRJNA507071 with Sequence Read Archive accession number SRR8256651. Supplemental material available at Figshare: https://figshare.com/s/49697abd3743f0d41b3a.
Results
X-linked C. briggsae introgressions lead to male sterility in C. nigoni but rescue the male sterility in hybrid F1 progeny
One of the common HIs between these two species is hybrid F1 male sterility between the two species with C. briggsae as the father (Figure 1). We previously identified at least two independent C. briggsae genomic fragments from its X chromosome that produced male sterility in C. nigoni as an introgression (Figure 2A) (Bi et al. 2015). However, the C. nigoni females that carry the same introgression are fertile. To systematically search for any C. briggsae genomic fragment that could rescue the inviability of F1(n, b) males, we performed reciprocal crosses between wild-type C. briggsae and C. nigoni lines that each carry an independent X- or autosome-linked introgression (see Materials and Methods). To achieve maximal coverage of the C. briggsae genome, we selected a subset of introgressions that showed little or no overlap, which we had generated in a previous study (Bi et al. 2015). In addition, to narrow the interval responsible for the rescue, we included several introgressions that overlapped with those that were confirmed to be able to produce changes in F1 phenotypes. As a result, we performed crosses for a total of 29 introgression lines, covering 94.6% of the C. briggsae genome, with complete coverage of the X chromosome (Table S1).
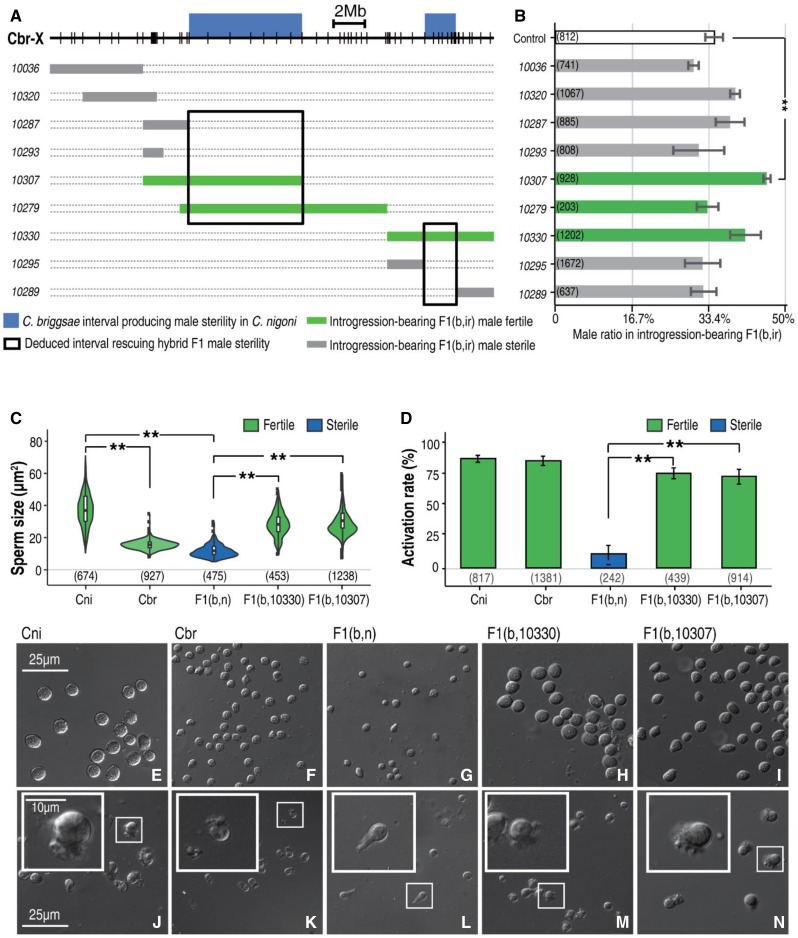
Figure 2.
Substitution of specific Cni X fragments with Cbr counterparts produces male sterility in Cni, but rescues male sterility in F1(b, n) hybrids. (A) Schematic representations of male fertility produced by Cbr introgressions in Cni or by hybrid F1(b, ir). Cbr X chromosome (Cbr-X) is shown as a black line. Positions for PCR primers used for genotyping are indicated as vertical lines (Table S2). Thick blue bars above the X chromosome indicate previously mapped Cbr chromosome intervals responsible for male sterility as an introgression in a Cni background. Thin bars underneath the X chromosome show the Cbr introgressions that produce hybrid F1(b, ir) fertile or sterile males, which are differentially color-coded in green and gray, respectively. For simplicity, the introgression names are shown as five-digit numbers on the left without the prefix “zzyIR.” Deduced intervals responsible for rescuing hybrid F1 male sterility are highlighted in black boxes. (B) Quantification of hybrid F1(b, ir) male ratio in all GFP-expressing F1 progeny shown in (A). Hybrid male fertility is color-coded as in (A). Note that none of the X-linked introgressions lead to a significant decrease in male ratio as opposed to the control except for zzyIR10307, which produces a significant increase (Student’s t-test, P < 0.05). Numbers of scored animals are indicated in parentheses. Control: male ratio in wild-type F1(b, n) hybrids (see also Figure S5, where all autosome-linked introgressions produce a significant decrease in male ratio). (C) Quantification of sperm sizes. Shown are violin plots of sperm sizes from the parental (Cni and Cbr) males, their hybrid F1(b, n) male, and the introgression-bearing F1 hybrid male between a wild-type C. briggsae father and C. nigoni mother carrying either of the two introgressions, i.e., zzyIR10330 and zzyIR10307, with fertility color-coded in dark blue (sterile) or green (fertile). Number of sperm scored is indicated. Note the size differences between wild-type Cbr and Cni males (P < 2.2e−16), and between the sterile and fertile hybrid males (P < 2.2e−16) (Wilcoxon signed-rank test). (D) Quantification of sperm activation rate. Shown are bar plots of activation rate from the males in (C). Rescued fertile hybrid males carrying introgressions demonstrate activation rates comparable to their parental species, which are significantly higher than those of the hybrid sterile F1(b, n) males (P < 0.01, two-sample Student’s t-test, unpaired). (E–N) Sperm morphology and activation in parental or hybrid males. Shown are the sperm before (E–I) and after activation (J–N). From left to right: parental (Cni and Cbr) males, their hybrid F1(b, n) male, and the introgression-bearing hybrid F1 male between a C. briggsae father and C. nigoni mother carrying either of the two introgressions, i.e., zzyIR10330 and zzyIR10307. Sperm after activation by pronase treatment are shown at the bottom. Magnified views are shown in insets. The morphology of activation (pseudopod) seems abnormal in the sterile F1 hybrid. Cbr, C. briggsae; Cni, C. nigoni.
Consistent with our hypothesis, we identified C. briggsae introgressions that could rescue hybrid F1 male fertility. Specifically, a cross between C. briggsae males and C. nigoni females carrying the introgressions produced hybrid fertile males, but a similar cross without the introgressions produced hybrid sterile males (Figure 1 and Figure 2A). The results suggest that there is a locus in the introgression that, when homozygous, interacts with another C. nigoni locus elsewhere in the genome, which is required for sterility rescue. The gonad morphology of the rescued males was similar to that of their parental males (Figure S1). The availability of a hybrid F1(b, n) fertile male opens the door for further genetic dissection of the X–autosome interaction that is required for hybrid male fertility as described below.
In hybrid F1 adult progeny fathered by C. briggsae, the hybrid F1 males, which were referred to as F1(b, n) males, were not only sterile but also demonstrated a disproportional segregation of sex ratio; that is, ∼35% of the hybrid F1 progeny were male and the rest were females, indicating that there are some other epistatic interactions that lead to hybrid male inviability. To investigate whether the mapped genomic interval that rescued the hybrid male sterility also rescued the hybrid male inviability, we quantified the ratio of introgression-bearing males and females among all the hybrid F1 progeny from the following cross, that is, between a C. briggsae male and a C. nigoni female carrying an introgression from various parts of the C. briggsae X chromosome. We found that only one of the X-linked introgressions rescued the hybrid male inviability (Figure 2B), suggesting that some loci affect fertility but not viability.
Sperm sizes of the rescued hybrid males are more similar to those of wild-type C. nigoni males
The sperm size of hermaphroditic C. briggsae is significantly smaller than that of dioecious C. nigoni (Vielle et al. 2016), and mating between the two species with C. nigoni as the father sterilized the maternal C. briggsae (Ting et al. 2014). We wondered how parental alleles controlled sperm sizes in the hybrid sterile males and in the F1 males with rescued fertility. Therefore, we quantified the sperm sizes derived from the hybrid sterile, rescued fertile, and parental males. We found that the sperm sizes of the hybrid F1(b, n) sterile males carrying a C. nigoni X and heterozygous autosomes were comparable to that of C. briggsae, suggesting that C. briggsae autosome alleles controlling sperm size are dominant. However, the sperm of the rescued hybrid fertile males, who carry the same genome as F1(b, n) males except for an X-linked introgression from C. briggsae, were significantly larger (Figure 2, C and E–I), arguing against the dominance of the C. briggsae autosomal alleles in controlling sperm size. We speculate that the small sperm size of sterile F1(b, n) males could be a secondary phenotype resulting from its underdevelopment or death, while the large sperm size of the hybrid males with rescued fertility may reflect the healthy development of vigorous cells. The results also suggested that spermatogenesis fails during the formation of primary spermatocytes in the hybrid sterile males (Vielle et al. 2016). Sperm activation rates were comparable between two rescued males, but were significantly higher than the hybrid F1(b, n) sterile males (Figure 2, D and J–N).
Few improvements on B2 breakdown when backcrossed to C. briggsae
One of prominent asymmetric HI phenotypes between C. briggsae and C. nigoni is the complete B2 breakdown of all progeny (no viable progeny) derived from crossing of the hybrid F1 female with a wild-type C. briggsae male, but not a C. nigoni male (Woodruff et al. 2010). The availability of hybrid F1(b, n) fertile males carrying zzyIR10330 or zzyIR10307 provided an opportunity to perform a reciprocal cross between C. briggsae and hybrid F1 progeny, allowing the evaluation of whether the B2 breakdown resulting from backcrossing to C. briggsae still holds with an increased contribution from the C. briggsae genome. To test this hypothesis, we scored the brood sizes, survival rates, and male ratios in the progeny derived from the crosses between the rescued F1(b, ir) males that carry the X-linked introgressions zzyIR10330 and zzyIR10307, and the sperm-depleted C. briggsae hermaphrodite (Figure 3). About one-quarter of ∼50 laid B2 embryos from the cross developed into adulthood (Figure 3, A and B). Notably, the male ratio was significantly higher than that in the F1(b, n) progeny (Figure 3C). However, all the surviving B2 males and females became sterile with deformed gonads (Figure S2), which suggests that the B2 inviability involved loci other than the X-linked introgressions used here.
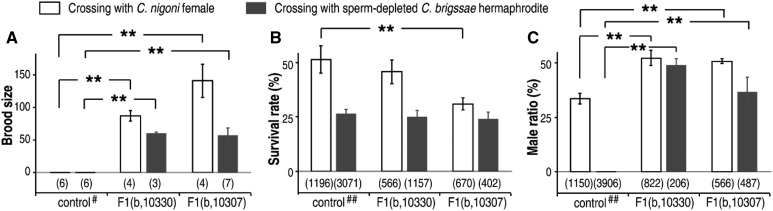
Figure 3.
Quantification of fertility rescue in hybrid F1 males. Shown are bar plots of brood sizes (A), survival rates (B), and male ratios (C) of hybrid B2 progeny between the rescued F1 fertile males [F1(b, 10330) or F1(b, 10307)] and sperm-depleted C. briggsae hermaphrodites or C. nigoni females, respectively. Numbers of the scored F1 males, B2 embryos, and adults are indicated underneath each plot in (A–C). Significant deviation from control is indicated by “**” (P < 0.01, Student’s t-test). #, brood size from crosses between F1(b, n) males and C. nigoni females (white box)/C. briggsae hermaphrodites (black box); ##, survival rates and male ratios of F1(b, ir) and F1(ir, b) progeny.
We also scored the survival rates in the progeny derived from the crosses in the opposite direction for all introgressions in this study, that is, the crosses between C. briggsae males and the F1 progeny, derived from reciprocal crosses between C. briggsae and C. nigoni carrying an autosome- or X-linked introgression. We were intrigued to find that few progeny survived to adulthood (Figure S3, A and B). Taken together, the increased contribution of the C. briggsae genome in the hybrid did not improve the fertility of the hybrid B2 progeny, thus preventing further backcrossing with C. briggsae beyond the B2 generation. Given that the fertility-rescued hybrid F1 males open up the possibility of an F1 × F1 intercross, we performed a cross between the fertility-rescued hybrid F1 males and F1 females, and only a small portion of progeny were able to survive to adulthood (Figure 3C). However, most of them looked very sick, suggesting too many combinations of HIs that severely damaged fitness. Further intercrosses of the surviving males and females produced no embryos.
A specific interaction between C. nigoni chromosome II and the C. briggsae X chromosome is required for hybrid male sterility in C. nigoni
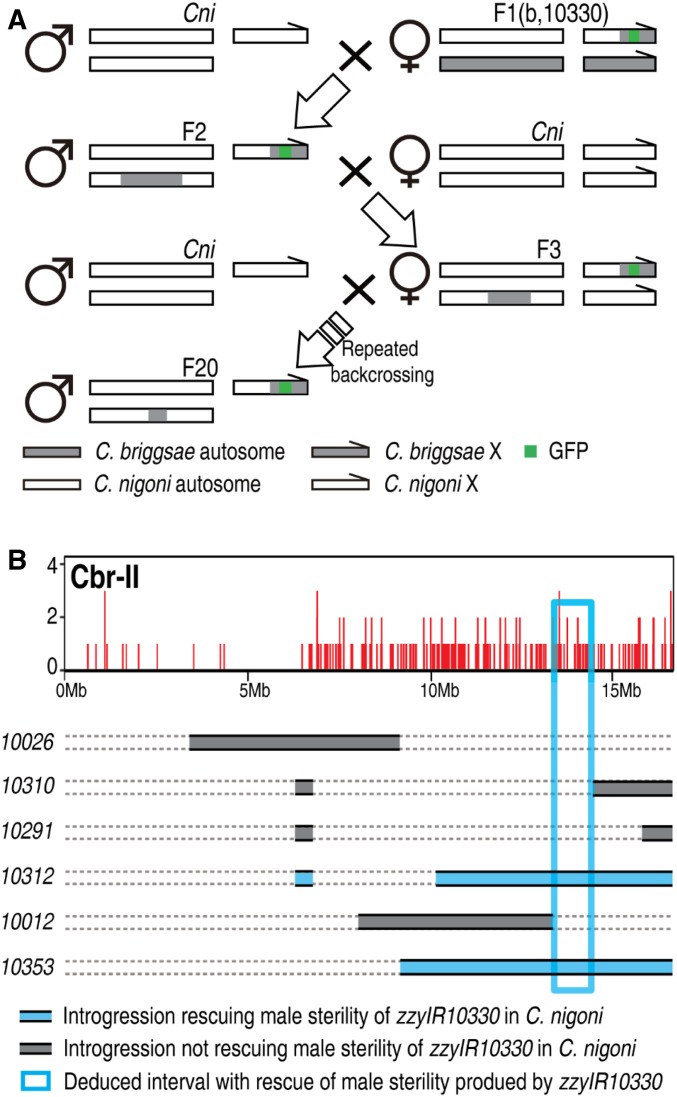
The availability of the hybrid F1 fertile male provided us with an opportunity to screen for specific X–autosome interactions that are required for hybrid male sterility (Figure 4A). Because the hybrid F1(b, n) males that carry either zzyIR10330 or zzyIR10307 are fertile, we randomly chose one of them, i.e., zzyIR10330, to isolate the autosomal interacting loci that are required for the observed rescue. We initiated the crosses by mating wild-type C. briggsae males and C. nigoni (zzyIR10330) females. The resulting introgression-bearing hybrid F1(b, 10330) (see Materials and Methods for nomenclature) fertile males were backcrossed with wild-type C. nigoni females for as many as 20 generations, using the introgression-bearing fertile male. Therefore, any C. briggsae loci that cosegregated with zzyIR10330 were expected to be identified by genotyping of the fertile males. Given the possibility of multiple independent C. briggsae loci that could cosegregate with the introgression and together rescue male sterility, we selected a single male to set up a cross for genotyping by single-worm PCR using the C. briggsae-specific primers described in Bi et al. (2015). We found that C. briggsae chromosome II had specifically cosegregated with zzyIR10330 (Figure 4B).
Figure 4.
Identification of C. briggsae (Cbr) autosomal loci that interact with introgression zzyIR10330 to rescue the sterility of introgression-bearing C. nigoni (Cni) males. (A) Backcrossing strategy. Hybrid fertile F1(b, 10330) male was backcrossed with wild-type C. nigoni for ≥ 20 generations. In addition to the male fertility check, introgression-bearing males were genotyped every three to four generations by PCR to ensure the absence of size changes to the introgression, zzyIR10330, which could lead to the rescue of male sterility. (B) Mapping of autosomal loci cosegregating with zzyIR10330. Top: genotyping results using next-generation sequencing (see also Figure S4), showing that the right arm of chromosome II is cosegregating with zzyIR10330, which is required for male fertility. Bottom: fine mapping of interacting loci on chromosome II using existing introgressions on chromosome II as indicated. The deduced smallest interval that rescues male sterility is highlighted in the rectangular box.
Given the error-prone features and limited resolution of the PCR-based method, it is possible that there were some other cosegregating segments that were missed by the PCR assay. To detect any other possible fragments that could cosegregate with zzyIR10330, we performed whole-genome sequencing with Nanopore technology using DNA extracted from the female progeny derived from the cross between introgression-bearing males and wild-type C. nigoni females (see Materials and Methods). These female progeny, but not the male progeny, were expected to carry the introgression and any cosegregating loci as heterozygotes in an essentially C. nigoni background. Consistent with the PCR-based assay, we detected only a single C. briggsae fragment on the right arm of chromosome II that cosegregated with zzyIR10330 in the hybrid rescued males (Figure 4B and Figure S4). The sequencing results also confirmed that the rescued male fertility did not result from a decreased length of zzyIR10330, but from the cosegregation of zzyIR10230.
The identified cosegregating zzyIR10230 introgression is more than one-half the size of chromosome II (Figure 4B), which was expected to result in inviability when rendered homozygous in C. nigoni based on our previous introgression results (Bi et al. 2015). To narrow the genomic interval responsible for the rescue of male sterility, we took advantage of our existing overlapping introgressions in the same region (Figure 4B). Specifically, we set up crosses between C. nigoni females that carried zzyIR10330 and six individual C. nigoni strains that each carried an independent introgression that overlapped with zzyIR10230. The presence and absence of fertile males would be indicative of the introgression’s ability or inability, respectively, to rescue the male sterility caused by zzyIR10330 in C. nigoni. Our cross results not only confirmed the significance of zzyIR10230 in the rescue of hybrid male sterility, but also allowed us to define an interval of 1.1 Mb from 13.35 Mb to 14.45 Mb on the C. briggsae Chromosome II (cb4), which interacts with zzyIR10330 to rescue the male sterility. Notably, the C. nigoni strain carrying the double introgression can be perpetuated as a stable line if needed by selecting for the lack of selfing and ongoing selection for male fertility, providing an opportunity for further downstream analysis.
Homozygosity of independent autosome introgressions rescues hybrid inviability of F1(n, b) males
Another prominent HI phenotype between C. briggsae and C. nigoni is the hybrid inviability of F1(n, b) males (Figure 1). Despite the rescue of hybrid F1(b, n) male sterility by the X-linked introgressions, these introgressions failed to rescue the inviability of the hybrid F1(n, b) males, indicating that differential epistatic interactions are responsible for parent-of-origin hybrid male fertility and viability.
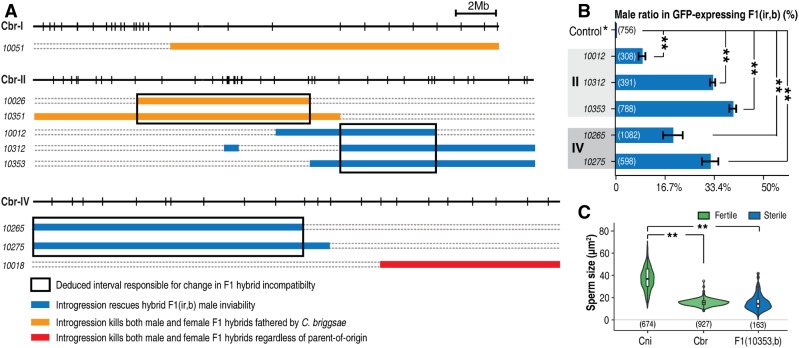
We were able to identify two independent autosomal intervals on the right and left arms of chromosomes II and IV, respectively, that rescue F1(n, b) male inviability (Figure 5A). However, the rescued males were sterile, and had malformed gonads and spontaneous sperm activation (Figure S5), which suggests that additional loci may be found elsewhere in the C. briggsae genome that are required for male fertility. Contrasting the overlapping parts of the relevant introgressions allowed us to narrow the intervals responsible for the observed rescue to roughly 3.8 and 8.9 Mb on chromosomes II and IV, respectively (Figure 5A). To examine whether the viability rescue was complete, we scored the ratio of introgression-bearing males among all introgression-bearing progeny. We found that the ratio of the introgression-bearing males was significantly smaller than the expected 50% for all of the five scored introgressions (Figure 5B), suggesting that the rescue of viability was incomplete.
Figure 5.
Deviated hybrid F1 phenotypes from those of crossing progeny between parental species in the presence of autosomal introgression. (A) List of deviated phenotypes in the hybrid F1 progeny. Chromosomes and introgressions are indicated as in Figure 2A. Introgressions with directional and unidirectional killing of hybrid F1 progeny (both male and female) are colored in orange and red, respectively. Introgressions that rescue hybrid male inviability in F1(n, b) are colored in blue. Deduced smallest intervals responsible for given phenotypes are highlighted by rectangular boxes. (B) Quantification of male ratio in introgression-bearing F1(ir, b) progeny with rescued male inviability (blue). Numbers of scored animals are indicated. Only introgressions on chromosomes II and IV are found to be able to rescue male inviability in F1(n, b) worms. The male ratio in wild-type F1(n, b) worms was scored as the control group. (C) Statistics of sperm sizes in parental or hybrid males. Shown are violin plots of sperm sizes of wild-type C. nigoni (Cni) or C. briggsae (Cbr) males, sterile F1 hybrids (b, n), and F1(10353, b) males with the number of scored sperm indicated below. Note that the sperm sizes of the rescued sterile males are comparable to those of C. briggsae, but are significantly smaller than those of C. nigoni males (Wilcoxon test, P < 2.2e−16) (Figure 2C).
To investigate the inheritance of sperm size in the rescued hybrid F1(n, b) males that carry a complete C. briggsae X chromosome, we quantified sperm sizes for one line of the viable males rescued by the introgression zzyIR10353. Again, we found that the sperm sizes of the rescued viable yet sterile males were comparable to those of wild-type C. briggsae males, but were significantly smaller than those of wild-type C. nigoni males (Figure 5C). These results demonstrate that a sperm is functional in the hybrid only when its size is comparable to that of wild-type C. nigoni males. They also suggest that a part of the C. briggsae X chromosome other than zzyIR10330 is incompatible with C. nigoni autosomes, which likely explains the observed male sterility.
Parent-of-origin-dependent or -independent killing of both male and female F1 progeny by different autosomal introgressions
Despite the sterility or the inviability of the hybrid F1 males, the hybrid F1 females resulting from crosses between C. briggsae and C. nigoni were mostly viable and fertile, regardless of their parent-of-origin (Figure 1) (Woodruff et al. 2010). To demonstrate whether there was any HI in hybrid F1 females, we performed crosses between wild-type C. briggsae males and C. nigoni females carrying an introgression derived from C. briggsae autosomes, and crosses in the opposite direction. It was expected that the introgression locus would be homozygous in the hybrid F1 progeny (Figure 1). Unexpectedly, we observed three independent autosomal intervals (deduced from overlapping introgressions if more than one introgressions producing the same phenotype were available) located on chromosomes I, II, and IV, respectively, which produced inviability of both hybrid male and female progeny that carried an introgression, hereafter termed complete inviability, when present as homozygotes achieved by crossing C. briggsae males and C. nigoni females carrying the introgression (Figure 5A). The intervals on chromosomes I and II were supported by one and two introgressions, respectively, which resulted in the complete inviability of F1(b, ir) worms. In contrast, the interval on chromosome IV was supported by a single introgression, which produced complete inviability that was independent of the parent-of-origin, meaning that complete inviability was observed in reciprocal crosses between wild-type C. briggsae and C. nigoni females carrying the autosomal introgression. Apparently, only nuclear genomes were involved in the inviability because the viability was not improved when C. nigoni mitochondria were substituted by their C. briggsae equivalents (Table S3). Given that the three introgressions produced no inviability in the otherwise C. nigoni background as a heterozygote, their epistatic interactions with the rest of the C. nigoni genome are likely to be recessive in the hybrid F1 background. It is also possible that the inviability may be due to a maternal vs. zygotic conflict. That is, the nuclear genome of the C. nigoni female carrying the introgression may encode a component that creates a lethal interaction with a zygotic gene product of C. briggsae in the F1 hybrid. No introgression on the X chromosome produced complete inviability when present in homozygotes or hemizygotes, indicating that the X chromosome is barely involved in hybrid female viability either in hybrid F1 progeny or in the C. nigoni strains that carry a C. briggsae introgression.
To evaluate the effects of autosomal introgression on male viability in hybrid F1 progeny, we scored the ratio of introgression-bearing males among all the hybrid F1 progeny that carried an introgression from crosses between C. briggsae males and C. nigoni females that carried an introgression. We found that the ratio of males was significantly lower than that of females in nearly all the hybrid progeny that carried an autosome-linked introgression (Figure S6). In contrast, the ratio of males was comparable to that of females in nearly all hybrid progeny that carried an X-linked introgression (Figure 1B), which is consistent with the observation that male-specific genes are enriched on autosomes but depleted on the X chromosome (Sturgill et al. 2007), although the idea was challenged in other species (Meiklejohn and Presgraves 2012).
Partial homozygosity for C. briggsae alleles rarely increased the incidence of selfing relative to the F1 generation
Another purpose of the crosses using the introgression strains was to explore whether the increased proportion of the C. briggsae genome in the hybrid could recover the selfing capability of the hybrid females. To this end, we examined the fertility of all crossed females that carried an introgression by plating a single hybrid L4 C. briggsae or C. nigoni female. We found that only the L4 progeny from the cross between a C. briggsae male and a C. nigoni female carrying introgression zzyIR10320 were able to produce ∼50 eggs without further mating. A couple of the eggs hatched and grew to adulthood. However, all of them were all sterile females, indicating that the increased proportion of C. briggsae genome in the hybrid did not result in the recovery of the selfing capability.
Discussion
Asymmetric male sterility or inviability is common in cross-species hybrids, but the mechanism responsible for these phenotypes is poorly defined from both genetic and molecular standpoints. Despite intensive study of hybrid male sterility or inviability, the mechanism of hybrid female inviability remains largely unknown. In this study, we investigated the genetic mechanism behind asymmetric defects in the viability and fertility of hybrid male and female progeny, resulting from crosses between hermaphroditic C. briggsae and dioecious C. nigoni.
Differential genetic mechanism between hybrid male sterility and viability, and female viability
The availability of hybrid F1(b, n) fertile males that carry a GFP-labeled introgression allowed us to dissect the genetic loci involved in hybrid F1 male sterility. By contrasting the genotypes between F1(b, n) sterile and F1(b, 10330) or F1(b, 10307) fertile males, we were able to deduce at least two independent incompatible interactions between the C. nigoni homologous region of zzyIR10330 or zzyIR10307 and the C. briggsae autosome in the hybrid F1(b, n) males, which are required for the observed sterility (Figure 6). This is because the only difference in genotype between the two was the presence of zzyIR10330 or zzyIR10307 in the fertile males, but not in the sterile males. This finding is consistent with the notion that the evolution of sex chromosomes is no longer dominated by the unique genetic features of the sex chromosomes themselves, but is instead a result of global interactions between sex chromosomes and autosomes (Lynch and Force 2000; Moyle et al. 2010; Long et al. 2012). The study of gene birth in the Drosophila genome has revealed disproportional translocation of X-linked genes to autosomes, which are predominately testis-specific genes (Bai et al. 2007). Given that the size of the hermaphroditic C. briggsae genome is substantially smaller than that of dioecious C. nigoni (Ren et al. 2018; Yin et al. 2018) and other Caenorhabditis species (Fierst et al. 2015), it remains to be determined whether differential gene translocation exists between the X chromosome and the autosomes between the two species that is responsible for the observed X–autosome interaction.
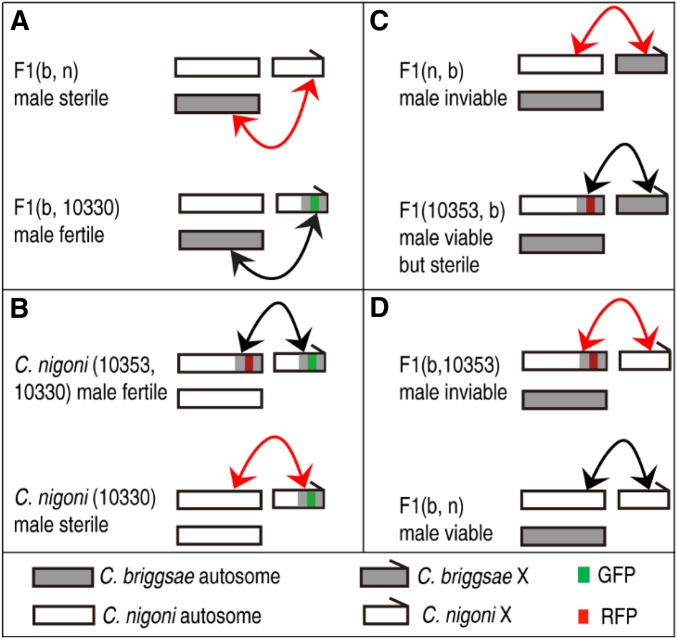
Figure 6.
Summary of interactions between X chromosome and autosomes in determining male fertility and viability. Shown are interactions using X-linked (zzyIR10330) and chromosome II-linked (zzyIR10353) introgressions as examples. The two introgressions are indicated as GFP and red fluorescent protein (RFP)-linked introgressions, respectively. Hybrid phenotypes are indicated. Compatible and incompatible interactions are indicated with black and red arrows, respectively. For simplicity, only chromosome II is indicated and introgression names are only denoted by their unique identifier within names. Note that only a single interaction is indicated, but it is possible that there are more interactions between other part of the autosomes and the X-linked fragment. (A) Hybrid F1(b, n) male sterility produced by an interaction between C. nigoni homozygous regions of zzyIR10330 and autosomes. (B) Hybrid male sterility of C. nigoni produced by an interaction between a small interval on C. nigoni chromosome II and zzyIR10330. (C) Hybrid F1 male inviability produced by an interaction between C. nigoni homozygous regions of zzyIR10353 and the C. briggsae X chromosome. (D) Hybrid F1(b, n) male inviability produced by an interaction between zzyIR10353 and the C. nigoni X chromosome.
F1(n, b) males are inviable (Figure 1). One of the two intervals that are required for the rescue of hybrid inviability in F1(n, b) males is located on C. briggsae chromosome II, which is the same as the one responsible for the rescue of sterility in C. nigoni males carrying zzyIR10330 (Figure 2A and Figure 5A). However, the region did not rescue the sterility of the F1(n, b) males (Figure 5, A and C), which indicates that hybrid male fertility and viability involve different loci. The other interval required for the rescue of the hybrid inviability of F1(n, b) males is located on chromosome IV. It is possible that the two intervals are more compatible with the C. briggsae X chromosome than its C. nigoni syntenic parts in the hybrid F1(n, b) background, which led to the rescue.
Intriguingly, all the X-linked introgressions that were viable and fertile as homozygotes/hemizygotes in C. nigoni worms were unable to rescue hybrid F1(b, n) male sterility, whereas those that produced hybrid male sterility as hemizygotes in a wild-type C. nigoni background were able to rescue male sterility (Figure 2A), which is consistent with the notion that multiple independent X–autosome interactions are involved in hybrid male sterility. It should be noted that our results do not preclude the possible role of a parental effect or the much smaller size of the X chromosome in C. briggsae than in C. nigoni being responsible for hybrid male sterility. Indeed, the parental effect has previously been blamed for observed hybrid inviability between C. remanei and C. laten (Bundus et al. 2018).
Rescued fertile F1(b, ir) males facilitate precise mapping of X–autosome interactions
Precise isolation of the interacting genomic interval is challenging in hybrid F1 animals due to complications caused by the coexistence of both parental genomes. However, backcrossing of the rescued F1(b, n) males with C. nigoni worms permits isolation of the precise interacting loci responsible for hybrid male fertility in C. nigoni (Figure 4A). We successfully isolated a C. briggsae interval that was 1.1 Mb in size on chromosome II that interacts with zzyIR10330, which is essential for hybrid male fertility in an otherwise C. nigoni background, indicating that a specific interaction between zzyIR10330 and the C. nigoni genomic interval homologous to the mapped C. briggsae interval is responsible for the C. nigoni male sterility (Figure 6). However, it remains possible that there are other autosomal loci that interact with the introgression and rescue male sterility, which may have been missed by our analysis. This possibility exists because we only randomly selected introgression-bearing males from each generation for backcrossing. Some other animals could have carried different introgressions other than the ones we isolated, which could rescue the sterility, but they could have been missed during the backcrosses used in this study. Given the relatively large sizes of the isolated genomic intervals involved in asymmetric hybrid male sterility and inviability, and female inviability, it is possible that any single genomic interval may contain more than one genetic locus responsible for the observed rescue or killing. Fine mapping of these loci remains a significant challenge. Future studies should focus on the development of more reagents to allow the systematic isolation of such interacting loci. One of the major limitations of our method of screening for interacting loci is the lack of introgressions labeled with differential markers, such as GFP and red fluorescent protein (RFP). RFP-labeled introgressions will allow direct testing of their interaction in C. nigoni via the crossing of two C. nigoni strains that each carry an independent autosomal introgression labeled by RFP and zzyIR10330 labeled by, for example, GFP. Rescue of male sterility by zzyIR10330 would be indicative of the two loci. It is worth noting that the genomic sequences of C. briggsae and C. nigoni diverge substantially from each other in both sequence identity and size (Ren et al. 2018; Yin et al. 2018), which significantly inhibits DNA recombination frequency. Selective loss of male-specific genes in hermaphroditic species and its subsequent reinforcement further complicates the precise mapping of HI loci using backcrossing. Therefore, the development of overlapping introgressions in different colors would provide an alternative to narrow the genomic interval for a given phenotype (Figure 4A).
Acknowledgments
We thank Chung Wai Shing and Cindy Tan for logistic support, and members of the Zhao laboratory for helpful comments. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). This work was supported by General Research Funds (grants HKBU12100118, HKBU12123716, and HKBU12100917) from the Hong Kong Research Grants Council, a Faculty Research grant, a Strategic Development Fund, and a Hong Kong Baptist University Interdisciplinary Research Cluster Fund (RC-IRCs/17-18/06) to Z.Z. The authors declare no competing interests.
Footnotes
Supplemental material available at Figshare: https://figshare.com/s/49697abd3743f0d41b3a.
Communicating editor: D. Presgraves
Literature Cited
- Bai Y., Casola C., Feschotte C., Betrán E., 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8: R11 10.1186/gb-2007-8-1-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. E., Sutherlin M. E., Emmons S. W., 1992. Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution 46: 585–594. 10.1111/j.1558-5646.1992.tb02067.x [DOI] [PubMed] [Google Scholar]
- Ben-David E., Burga A., Kruglyak L., 2017. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science 356: 1051–1055. 10.1126/science.aan0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y., Ren X., Yan C., Shao J., Xie D., et al. , 2015. A Genome-wide hybrid incompatibility landscape between Caenorhabditis briggsae and C. nigoni. PLoS Genet. 11: e1004993 10.1371/journal.pgen.1004993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundus J. D., Alaei R., Cutter A. D., 2015. Gametic selection, developmental trajectories, and extrinsic heterogeneity in Haldane’s rule. Evolution 69: 2005–2017. 10.1111/evo.12708 [DOI] [PubMed] [Google Scholar]
- Bundus J. D., Wang D., Cutter A. D., 2018. Genetic basis to hybrid inviability is more complex than hybrid male sterility in Caenorhabditis nematodes. Heredity 121: 169–182. 10.1038/s41437-018-0069-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., 1985. Genetic studies of three sibling species of Drosophila with relationship to theories of speciation. Genet. Res. 46: 169–192. 10.1017/S0016672300022643 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation, pp. 545 Oxford University Press, Oxford. [Google Scholar]
- Dey A., Chan C. K. W., Thomas C. G., Cutter A. D., 2013. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc. Natl. Acad. Sci. USA 110: 11056–11060. 10.1073/pnas.1303057110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Jin Q., Chen Y.-C., Cutter A. D., 2014. Gonad morphogenesis defects drive hybrid male sterility in asymmetric hybrid breakdown of Caenorhabditis nematodes. Evol. Dev. 16: 362–372. 10.1111/ede.12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., Braendle C., Cutter A. D., 2014. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One 9: e94723 [corrigenda: PLos One 10: e0118327 (2015)]. 10.1371/journal.pone.0094723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst, J. L., J. H. Willis, C. G. Thomas, W. Wang, R. M. Reynolds et al., 2015 Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet 11 e1005323 (erratum PLos Genet. 11 e1005497). 10.1371/journal.pgen.1005323 10.1371/journal.pgen.1005323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Andersen E. C., Shapiro J. A., Gerke J. P., Kruglyak L., 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578. 10.1126/science.1214318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K., Qadota H., Benian G. M., 2017. Development, structure, and maintenance of C. elegans body wall muscle, (April 13, 2017), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.81.2, http://www.wormbook.org. 10.1895/wormbook.1.81.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto N., Matsumoto K., 2017. Signal transduction cascades in axon regeneration: insights from C. elegans. Curr. Opin. Genet. Dev. 44: 54–60. 10.1016/j.gde.2017.01.010 [DOI] [PubMed] [Google Scholar]
- Huang R.-E., Ren X., Qiu Y., Zhao Z., 2014. Description of Caenorhabditis sinica sp. n. (Nematoda: Rhabditidae), a nematode species used in comparative biology for C. elegans. PLoS One 9: e110957 10.1371/journal.pone.0110957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K. C., Felix M. A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339 10.1186/1471-2148-11-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowska J. L., Ahmad A. R., Jahesh E., Cutter A. D., 2012. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evolution 66: 1180–1195. 10.1111/j.1558-5646.2011.01514.x [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., 1997. Dobzhansky’s genetics and the origin of species: is it still relevant? Genetics 147: 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, M., M. D. Vibranovski, and Y. E. Zhang, 2012 Evolutionary interactions between sex chromosomes and autosomes, pp. 101–114 in Rapidly Evolving Genes and Genetic Systems Oxford University Press, Oxford. 10.1093/acprof:oso/9780199642274.003.0011 [DOI] [Google Scholar]
- Lynch M., Force A. G., 2000. The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156: 590–605. 10.1086/316992 [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D. A., 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45: 331–355. 10.1146/annurev-genet-110410-132514 [DOI] [PubMed] [Google Scholar]
- Mango S. E., 2009. The molecular basis of organ formation: insights from the C. elegans foregut. Annu. Rev. Cell Dev. Biol. 25: 597–628. 10.1146/annurev.cellbio.24.110707.175411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly J. P., Presgraves D. C., 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5: e243 10.1371/journal.pbio.0050243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Presgraves D. C., 2012. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol. Evol. 4: 1007–1016. 10.1093/gbe/evs077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P. A., Ritchie M. G., Bailey N. W., 2017. A rare exception to Haldane’s rule: are X chromosomes key to hybrid incompatibilities? Heredity 118: 554–562. 10.1038/hdy.2016.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Muir C. D., Han M. V., Hahn M. W., 2010. The contribution of gene movement to the “two rules of speciation”. Evolution 64: 1541–1557. 10.1111/j.1558-5646.2010.00990.x [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nelson G. A., Ward S., 1980. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19: 457–464. 10.1016/0092-8674(80)90520-6 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343. 10.1016/j.tig.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragavapuram V., Hill E. E., Baird S. E., 2016. Suppression of F1 male-specific lethality in Caenorhabditis hybrids by cbr-him-8. G3 (Bethesda) 6: 623–629. 10.1534/g3.115.025320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Li R., Wei X., Bi Y., Ho V. W. S., et al. , 2018. Genomic basis of recombination suppression in the hybrid between Caenorhabditis briggsae and C. nigoni. Nucleic Acids Res. 46: 1295–1307. 10.1093/nar/gkx1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Koboldt D. C., Staisch J. E., Chamberlin H. M., Gupta B. P., et al. , 2011. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 7: e1002174 10.1371/journal.pgen.1002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., et al. , 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L. E., Haag E. S., 2017. Revisiting suppression of interspecies hybrid male lethality in Caenorhabditis nematodes. G3 (Bethesda) 7: 1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilthuizen M., Giesbers M. C. W. G., Beukeboom L. W., 2011. Haldane’s rule in the 21st century. Heredity 107: 95–102. 10.1038/hdy.2010.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. 10.1126/science.1151107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J., He K., Wang H., Ho W. S., Ren X., et al. , 2013. Collaborative regulation of development but independent control of metabolism by two epidermis-specific transcription factors in Caenorhabditis elegans. J. Biol. Chem. 288: 33411–33426. 10.1074/jbc.M113.487975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Zhang Y., Parisi M., Oliver B., 2007. Demasculinization of X chromosomes in the Drosophila genus. Nature 450: 238–241. 10.1038/nature06330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J. J., Woodruff G. C., Leung G., Shin N. R., Cutter A. D., et al. , 2014. Intense sperm-mediated sexual conflict promotes reproductive isolation in Caenorhabditis nematodes. PLoS Biol. 12: e1001915 10.1371/journal.pbio.1001915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Moyle L. C., 2007. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics 176: 1059–1088. 10.1534/genetics.106.065979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A., Callemeyn-Torre N., Gimond C., Poullet N., Gray J. C., et al. , 2016. Convergent evolution of sperm gigantism and the developmental origins of sperm size variability in Caenorhabditis nematodes. Evolution 70: 2485–2503. 10.1111/evo.13043 [DOI] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Felix M. A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012. 10.1534/genetics.110.120550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Bi Y., Yin D., Zhao Z., 2012. A method for rapid and simultaneous mapping of genetic loci and introgression sizes in nematode species. PLoS One 7: e43770 10.1371/journal.pone.0043770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Schwarz E. M., Thomas C. G., Felde R. L., Korf I. F., et al. , 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359: 55–61. 10.1126/science.aao0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Cook D. E., Zdraljevic S., Brady S. C., Lee D., et al. , 2018. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl. Trop. Dis. 12: e0006368 10.1371/journal.pntd.0006368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Boyle T. J., Bao Z., Murray J. I., Mericle B., et al. , 2008. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev. Biol. 314: 93–99. 10.1016/j.ydbio.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures and supplemental figures, and tables and supplemental tables. Table S1 lists the details of the introgression strains used in hybrid F1 phenotypic screening. Table S2 lists the newly designed genotyping primers based on C. briggsae genome assembly (cb4) in addition to those listed previously (Bi et al. 2015). Table S3 shows the statistics of hybrid progeny resulting from crosses that produced complete inviability. The Nanopore sequencing data were submitted to the National Center for Biotechnology Information under BioProject number PRJNA507071 with Sequence Read Archive accession number SRR8256651. Supplemental material available at Figshare: https://figshare.com/s/49697abd3743f0d41b3a.