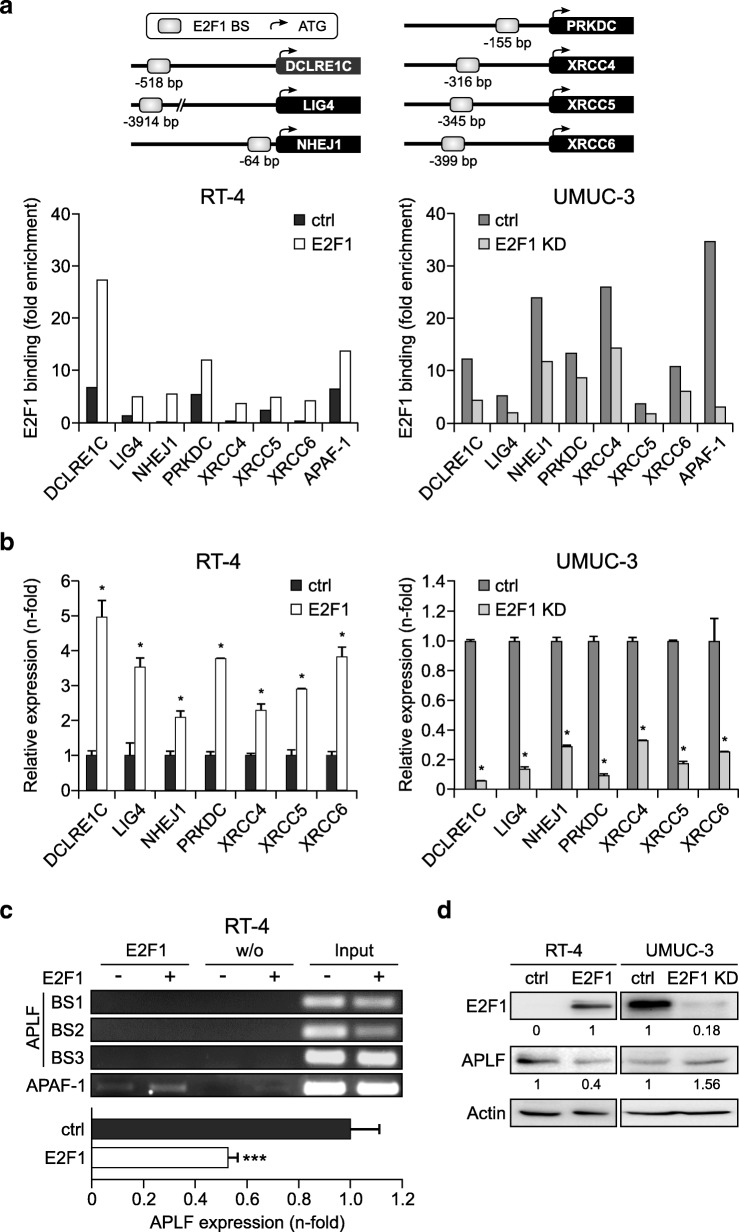
Fig. 2.
E2F1 transactivates c-NHEJ factors directly, but downregulates APLF indirectly. a Top: Schematic representation of the positions of the E2F1 responsive elements on the promoter of depicted c-NHEJ genes relative to the ATG codon. Primers were designed that encompass these sites, and the ability of E2F1 to directly interact with the corresponding elements in vivo was demonstrated by ChIP using specific E2F1 monoclonal antibody, followed by genomic PCR analysis. Bottom: ChIP assays, using anti-E2F1 antibody in RT-4 after ectopic E2F1 expression (left) and in stable UMUC-3 E2F1 knockdown cells (E2F1 KD) versus their controls (ctrl, right) demonstrated functional binding sites in the promoters of DCLRE1C, LIG4, NHEJ1, PRKDC, XRCC4, XRCC5, and XRCC6 genes. Mock IPs of the cell lysates were used as a background control and binding was expressed as fold-enrichment of each sample relative to mock IP controls. The E2F1 binding site of the APAF-1 promoter was used as a positive control for functional E2F1 binding sites. b qPCR mRNA levels of above-mentioned c-NHEJ factors in BC cell lines with (left) and without E2F1 (right). Asterisks indicate statistically significant (* p < 0.05) changes in comparison to the controls. c Top: ChIP assays in RT-4-E2F1 cells versus RT-4-ctrl for the APLF promoter regions − 673 to − 472 (BS1), − 492 to − 316 (BS2), and − 67 to + 75 (BS3) bps relative to the ATG codon showed no E2F1 binding in any of the three regions. Mock IPs incubated without antibody (w/o Ab) were used as a negative control, while input was used as positive control. The APAF-1 promoter was used as an E2F1 binding control. Bottom: qPCR in RT-4-E2F1 cells versus RT-4-ctrl cells for APLF (*** p < 0.001). d Detection of E2F1 and APLF protein levels in indicated BC cells. Band quantification was performed by imageJ software