Fig. 3.
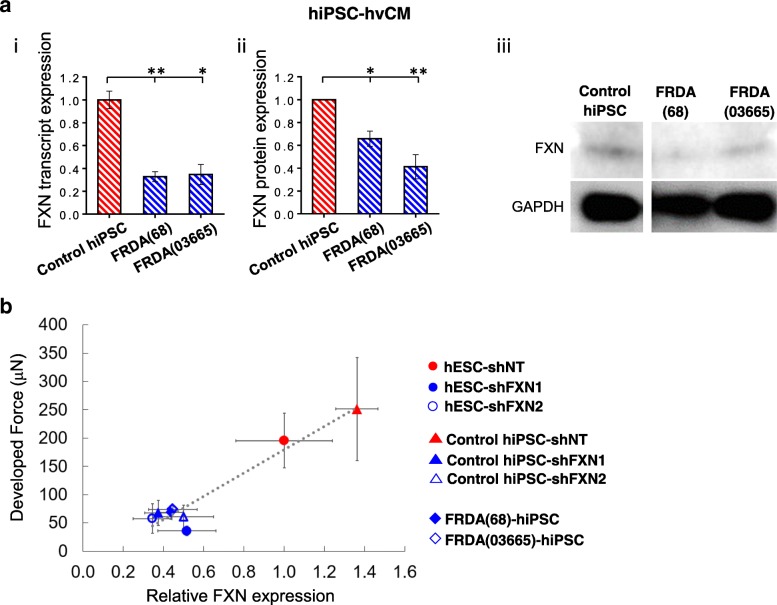
FRDA cardiac model derived from FRDA-hiPSCs. a FXN (i) transcript and (ii) protein expression (normalized to GAPDH expression) and (iii) representative Western blot image of FRDA(68)-hiPSC-hvCMs and FRDA(03665)-hiPSC-hvCMs relative to healthy control hiPSC-hvCMs (n = 3–5). Transcript and protein data were generated by 3–4 and 3–5 independent differentiations, respectively. b Correlation of developed force to FXN transcript expression in hESC- and hiPSC-hvCTS transduced with Lv-shNT, Lv-shFXN1, Lv-shFXN2, and FRDA-hiPSC-hvCTS relative to the hESC-shNT group (on day 12, at 1 Hz pacing). Force generation and FXN transcript expression data were generated from 3 to 8 and 3 to 6 independent cell differentiation batches, respectively. Data are shown as mean ± SEM. Statistical significance indicated by *p < 0.05 and **p < 0.01