Abstract
The envelope protein of Zika virus (ZIKV) exists as a dimer on the mature viral surface and is an attractive antiviral target because it mediates viral entry. However, recombinant soluble wild-type ZIKV envelope (wtZE) might preferentially exist as monomer (monZE). Recently, it has been shown that the A264C substitution could promote formation of dimeric ZIKV envelope protein (ZEA264C), requiring further characterization of purified ZEA264C for its potential applications in vaccine development. We also noted that ZEA264C, connected by disulfide bond, might be different from the noncovalent native envelope dimer on the virion surface. Because the antibody Fc fragment exists as dimer and is widely used for fusion protein construction, here we fused wtZE to human immunoglobulin G1 (IgG1) Fc fragment (ZE-Fc) for noncovalent wtZE dimerization. Using a multistep purification procedure, we separated dimeric ZEA264C and ZE-Fc, revealing that they both exhibit typical β-sheet–rich secondary structures and stabilities similar to those of monZE. The binding activities of monZE, ZEA264C, and ZE-Fc to neutralizing antibodies targeting different epitopes indicated that ZEA264C and ZE-Fc could better mimic the native dimeric status, especially in terms of the formation of tertiary and quaternary epitopes. Both ZEA264C and ZE-Fc recognize a ZIKV-sensitive cell line as does monZE, indicating that the two constructs are still functional. Furthermore, a murine immunization assay disclose that ZEA264C and ZE-Fc elicit more neutralizing antibody responses than monZE does. These results suggest that the two immunogen candidates ZEA264C and ZE-Fc have potential utility for neutralizing antibody selection and vaccine design against ZIKV.
Keywords: antibody, vaccine, flavivirus, dimerization, protein structure, dimer, envelope protein, Fc fragment, quaternary epitope, Zika virus, immunogen, neutralizing antibody, vaccine development, Flaviviridae, neurological disease
Introduction
Zika virus (ZIKV),2 as a re-emerging viral pathogen, belongs to the Flaviviridae family including dengue virus, West Nile virus, Japanese encephalitis virus, yellow fever virus, and tick-borne encephalitis virus (1, 2). It can be transmitted by Aedes mosquitoes and cause severe neurological diseases including Guillain-Barré syndrome in the adult (3, 4), and congenital Zika syndrome in the infant that includes microcephaly, brain abnormalities, and other severe birth defects (5–7). Because of the huge threat of ZIKV to the public health, it has raised worldwide attention and lots of work on the development of drugs and vaccines against ZIKV is in progress (8–10). However, there is no approved anti-ZIKV reagents for clinical use, which needs continuous efforts.
Like other flaviviruses, the genome of ZIKV encodes a single polyprotein which can be cleaved into three structural proteins (capsid, pre-membrane, and envelope (E)) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) (11–13). Among these proteins, E protein plays a very important role in viral entry (14–16). Therefore it is an effective target for inhibition of the virus. As a typical class II viral envelope protein, E proteins on the surface of mature ZIKV particle form antiparallel homodimers in a herringbone pattern (13, 17, 18). As for other flavivirus E proteins, the monomeric ZIKV E protein also has three distinct domains: a central β-barrel–shaped domain I (DI), an extended dimerization domain II (DII), and a C-terminal immunoglobulin-like domain III (DIII). The fusion loop (FL) of E protein is located in the distal end of DII and consists of hydrophobic residues that can insert into endosomal membranes during pH-dependent conformational changes and drive fusion (19).
Various monoclonal antibodies (mAbs) with different neutralizing activities against ZIKV have been identified that could bind to different epitopes in ZIKV E DI, DII, and DIII. For example, the antibodies that bind to DIII (e.g. ZV-67 and ZKA64) have high neutralizing activity without crossreactivity and might slightly cause antibody-dependent enhancement of infection (ADE) (20, 21), whereas those binding to FL epitope (FLE) (e.g. 2A10G6) are crossreactive but relatively modest in neutralizing, and tend to cause ADE (22, 23). Furthermore, a panel of antibodies recognizing quaternary epitope formed by E dimer (E-dimer–dependent epitope, EDE) were identified that are more potent and crossreactive, and have less ADE (21, 24–26). In addition, several neutralizing antibodies (e.g. Z3L1 and Z20) with strong neutralizing activity but no crossreactivity that target conformational epitope composed of residues in DI, DII, and DIII (tertiary epitope) were also identified (27). Therefore, preparation of dimeric E as its native status is a key point for development of effective immunogen for selection of powerful neutralizing antibodies and design of effective vaccines. However, expression of soluble WT ZIKV E protein (wtZE) might only lead to generation of monomeric E protein (monZE) (28, 29). Recently, it has been reported that introduction of a single Cys substitution (A259C) in E protein of dengue virus could lead to formation of dimeric E through an intersubunit disulfide bond (30). By the similar strategy, a ZIKV E protein mutant by replacement of Ala-264 to one Cys in ZIKV E protein (ZEA264C) was also designed to show that folding and dimerization of secretory ZIKV E proteins are strongly dependent on temperature (28). However, the conformational and functional information of covalent dimeric ZEA264C were still unclear and should be well-characterized for its potential use as an ideal immunogen. Because ZEA264C exists as covalent dimer connected by a disulfide bond, it also raises a question whether E dimer could form noncovalent linkage in vitro because native E dimer on virion surface is noncovalent. As a stable dimer, antibody Fc fragment has been widely used in construction of fusion protein for therapeutic purpose and vaccine design because it could make the fused protein bivalent (31). Hence, we proposed that wtZE could be noncovalently dimerized by fusing it with Fc fragment, and then also made Fc-fusion protein (ZE-Fc) as a candidate. Here, we combined different methods to structurally and functionally characterize monZE, ZEA264C, and ZE-Fc in vitro and in vivo. Our results disclose that major neutralizing epitopes including EDE were still maintained in ZEA264C and ZE-Fc. Moreover, immunization of these three proteins in mice shows that ZEA264C and ZE-Fc are more effective in eliciting antisera against ZIKV than monZE. Hence, both ZEA264C and ZE-Fc have potentials as promising immunogens for development of neutralizing antibodies including those target tertiary/quaternary epitopes, and potent vaccines.
Results
Design, expression, and separation of soluble ZIKV E dimer
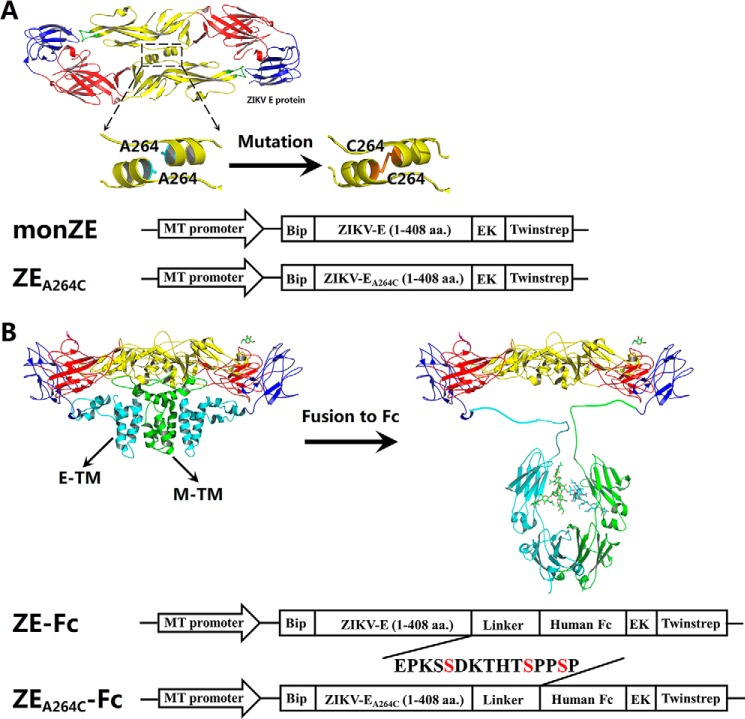
As reported previously, the ectodomain of ZIKV E (1–408 amino acids) (monZE) was used for soluble expression (32) as monomeric form (Fig. 1A). The residue Ala-264 in ZIKV E protein was mutated to Cys to generate ZEA264C, which was desired to form dimer (28) (Fig. 1A).
Figure 1.
Design and construction of monZE, ZEA264C, and Fc-fusion proteins for expression in Drosophila S2 cells. A, design of covalent E dimer. The A264C mutation was introduced according to the dimeric ZIKV E structure presented by PyMOL (PDB ID: 5LBV) (25) for generation of ZEA264C. The ectodomain (1–408 amino acids (aa)) of monZE and ZEA264C were placed between an N-terminal BiP secretion signal peptide and a C-terminal enterokinase cleavage site followed by Twin-strep-tag, under the control of an inducible Drosophila metallothionein (MT) promoter. B, design of ZIKV E and Fc-fusion proteins. According to the cryo-EM structure of ZIKV virion (PDB ID: 5IZ7) (17), E-TM interacts with M-TM to facilitate the formation of E dimer. Replacement of these regions by antibody Fc fragment (PDB ID: 1HZH) (35) might compensate for the loss of interaction and promote the formation of E dimer. The schematic diagram for design of expression of ZE-Fc and ZEA264C-Fc, as design of expression of monZE and ZEA264C, was also shown.
According to the cryo-EM structure of ZIKV virion (PDB ID: 5IZ7) (17), there are two transmembrane helices (E-TM) at the C terminus of one E protein that interacts with transmembrane domains of M protein (M-TM) (Fig. 1B), which has also been illustrated well in other flaviviruses (33, 34). The lacking of this interaction might result in the loss or significant reduction of dimerization of E protein. Therefore, we fused wtZE to antibody Fc fragment to construct a fusion protein ZE-Fc that was desired to form dimer noncovalently because of the strong interaction between two CH3 domains in Fc fragment (PDB ID: 1HZH) (35) (Fig. 1B). In addition, A264C in ZIKV E protein was also introduced in ZE-Fc (ZEA264C-Fc) to show whether the disulfide bond could enhance the dimerization of E protein when fused to Fc or not (Fig. 1B).
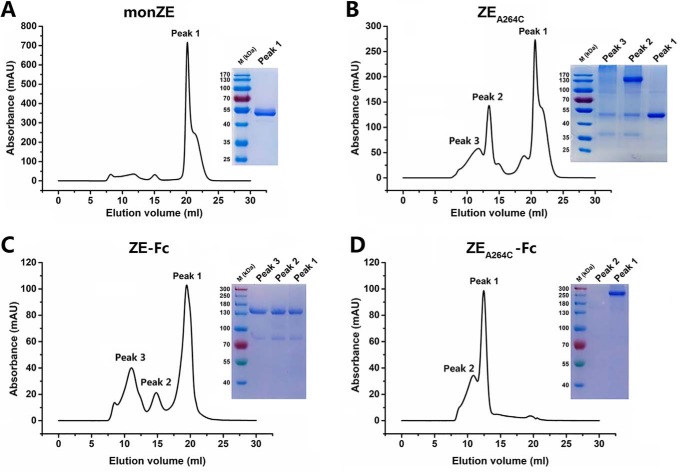
All four constructs (monZE, ZEA264C, ZE-Fc, and ZEA264C-Fc) were expressed in Drosophila S2 cells. After first-step purification by Strep-Tactin affinity chromatography column, the samples were subjected to size exclusion chromatography (SEC) for further separation. In the case of purified monZE, only one peak (Peak 1) was observed (Fig. 2A) whereas three major peaks (Peak 1, Peak 2, and Peak 3) were observed in the case of purified ZEA264C (Fig. 2B). According to analysis of three peaks by SDS-PAGE, it could be concluded that dimeric ZIKV EA264C mainly formed in Peak 2, whereas Peak 1 represented unpaired monomeric ZIKV EA264C, and Peak 3 indicated bigger oligomer. Therefore, we collected Peak 1 in the case of monZE and Peak 2 in the case of ZIKV EA264C for following experiments. For Fc-fusion proteins, we also checked all the peaks by SDS-PAGE. Although all of peaks of ZE-Fc could migrate to position of correct dimer according to the marker, only main peak (Peak 1) was collected (Fig. 2C). However, in the case of ZEA264C-Fc, Peak 1 was the main peak and the migration of it on SDS-PAGE indicated the formation of soluble aggregation (Fig. 2D). Taken together, monZE from Peak 1, ZEA264C from Peak 2, and ZE-Fc from Peak 1 were used for further analysis.
Figure 2.
Second-step purification with SEC. A, purification of monZE. Only one peak was separated from eluted monZE protein by SEC. The SDS-PAGE showed the molecular mass was correct as monomer. B, purification of ZEA264C. Three peaks were separated from eluted ZEA264C protein by SEC. Dimer only formed in Peak 2 analyzed by SDS-PAGE under nonreducing condition. C, purification of ZE-Fc. Three peaks were observed in eluted ZE-Fc. Each peak was loaded on SDS-PAGE without boiling. Although all of them may exist as dimer according to the migration, only the main peak (Peak 1) was collected. D, purification of ZEA264C-Fc. Two peaks were separated from eluted ZEA264C-Fc. Each peak was loaded on SDS-PAGE without boiling, which indicated that the main peak (Peak 1) existed as soluble aggregates.
Formation of dimer
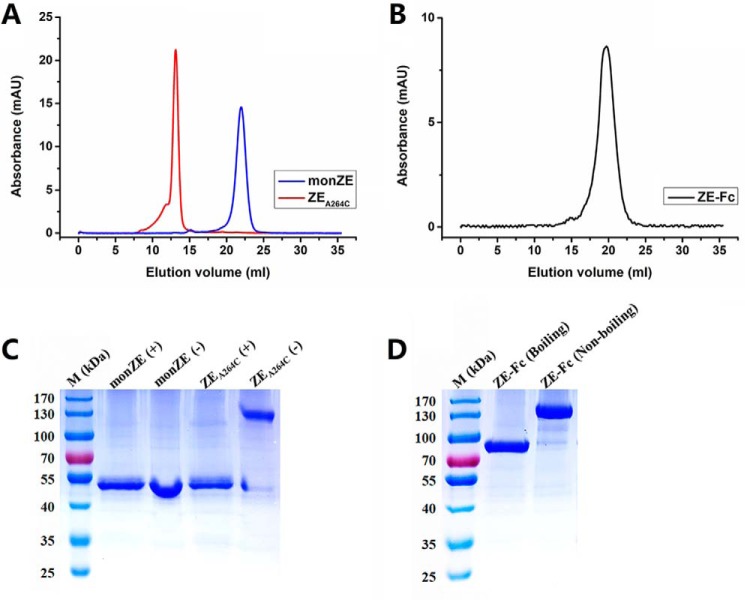
After concentration, the purified monZE, ZEA264C, and ZE-Fc were run on SEC again and no additional peak was observed, which indicated that they were in stable status in solution (Fig. 3, A and B). Then all of them were confirmed on SDS-PAGE under different conditions. By comparison of the migration in nonreduction and reduction conditions, it could clearly find that ZEA264C forms dimer through disulfide bridge (Fig. 3C). Similarly, ZE-Fc could also form dimer, which was deduced from its migration in boiling and nonboiling conditions (Fig. 3D). To confirm molecular mass (M.M.) of ZEA264C that should be double mass of monZE, we also performed the MALDI-TOF MS assay. The M.M. values from the assay were 50.0 kDa for monZE and 100.1 kDa for ZEA264C, which matched the theoretical M.M. values of E monomer (48.6 kDa) and dimer (97.2 kDa) well. In conclusion, both ZEA264C and ZE-Fc form stable dimer in solution.
Figure 3.
Molecular mass of purified monZE, ZEA264C, and ZE-Fc. A and B, SEC evaluation. Only one unique peak in each protein was observed, indicating they were uniform. C and D, SDS-PAGE analysis. monZE and ZEA264C were loaded in the presence (+) or absence (−) of reducing agent DTT; ZE-Fc was loaded after boiling or not.
Secondary structure and stability
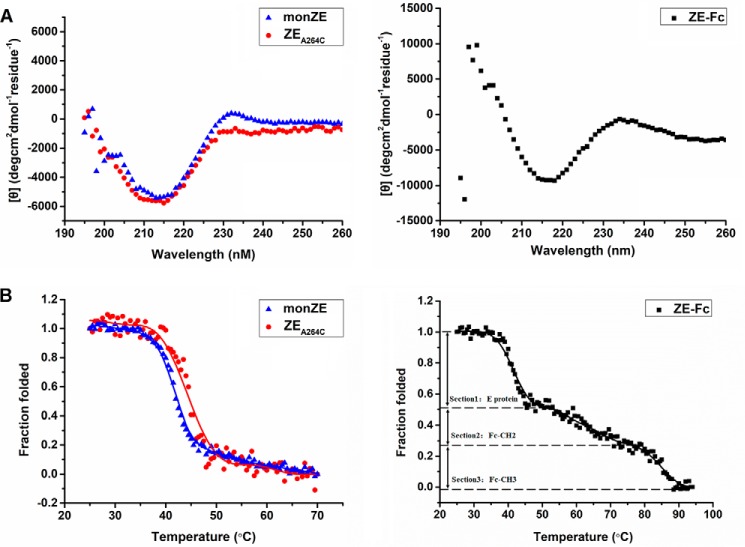
Circular dichroism (CD) spectra of these three proteins exhibited a maximum negative peak between 216 and 218 nm, illustrating that these proteins adopted rich β-sheet secondary structures (Fig. 4A). The highly overlapping structural profiles showed that introduced Cys mutation does not significantly change the overall structure (ZEA264C versus monZE). As the temperature increased, the structure was destroyed with obviously S-shaped curve (two-state) in the case of monZE and ZEA264C (Fig. 4B). The melting temperature (Tm) values of both proteins were calculated by Boltzman fitting equation (Tm of monZE: 42.1 °C and Tm of ZEA264C: 44.5 °C). Although it seemed that ZEA264C was slightly more thermostable, the dimerization might not obviously alter the stability of each domain in E protein. These results demonstrated the successful formation of dimeric ZIKV E protein by engineered disulfide bond without change of overall structure. The maximum negative peak in the case of ZE-Fc located between 216 and 218 nm as desired because Fc is also mainly composed of β-strands (Fig. 4A). The thermo-induced unfolding curve of ZE-Fc exhibited three sections including unfolding of E protein and CH2 and CH3 domains in Fc fragment. The unfolding process of E protein in Fc-fusion protein was quite similar to that of monZE/ZEA264C, whereas the unfolding of CH2 and CH3 domains in Fc fragment were similar to our previous result (Fig. 4B) (36). Therefore, it is reasonable to believe that fusion of wtZE with Fc has no obvious influence on secondary structure of E protein.
Figure 4.
Secondary structure and thermo-induced unfolding curve measured by CD. A, secondary structure at 25 °C. A major negative peak at 216 nm was observed in monZE, ZEA264C, and ZE-Fc, which indicated a typical β-sheet structure. B, Thermo-induced unfolding curve was plotted by recoding the CD signals when temperature was climbing. The unfolding curve of ZE-Fc was separated into three sections: Section 1, E protein in ZE-Fc protein; Section 2, CH2 domain in Fc (Fc-CH2); and Section 3, CH3 domain in Fc (Fc-CH3).
Recognition by neutralizing antibodies targeting nonquaternary epitopes
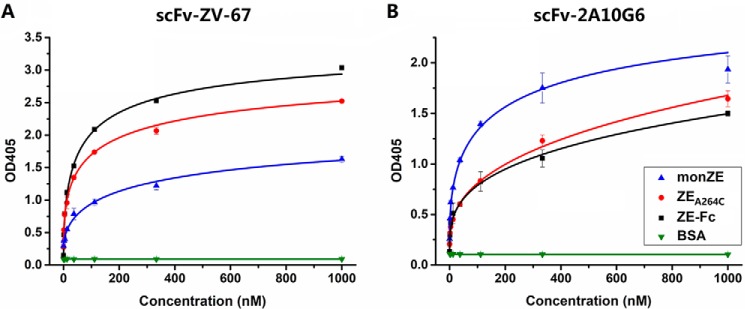
The ability of these three proteins to be recognized by neutralizing antibodies scFv-ZV-67 (targeting DIII) and scFv-2A10G6 (targeting FL) was firstly tested by ELISA. The EC50 values of binding of scFv-ZV-67 to ZEA264C and ZE-Fc were 145 and 68 nm, respectively, whereas the value was 405 nm in the cases of monZE (Fig. 5A). In contrast, scFv-2A10G6 bound to ZEA264C and ZE-Fc with EC50 values of 170 and 114 nm, whereas the value decreased to 65 nm in the cases of wtZE (Fig. 5B). In comparison of these two sets of results, the bindings of 2A10G6 to ZEA264C and ZE-Fc were relatively reduced, possibly because of burying of the fusion loop region after dimerization. These observations provide the possibility that dimeric E protein would elicit fewer antibodies targeting FLE as immunogen than does monomeric E, and therefore reduced ADE could be desired (30).
Figure 5.
Recognition of DIII and FLE on E proteins. A and B, binding of scFv-ZV-67 (targeting DIII) (A) and scFv-2A10G6 (targeting FLE) (B) to monZE, ZEA264C, and ZE-Fc. The monZE, ZEA264C, ZE-Fc, and BSA were coated on plates and serially diluted positive antibodies were added to test the binding respectively. HRP-conjugated anti-His antibody was used as second antibody in both experiments. Error bar, the data are shown as mean ± S.D. from two independent experiments.
Recognition by neutralizing antibodies targeting tertiary/quaternary epitopes
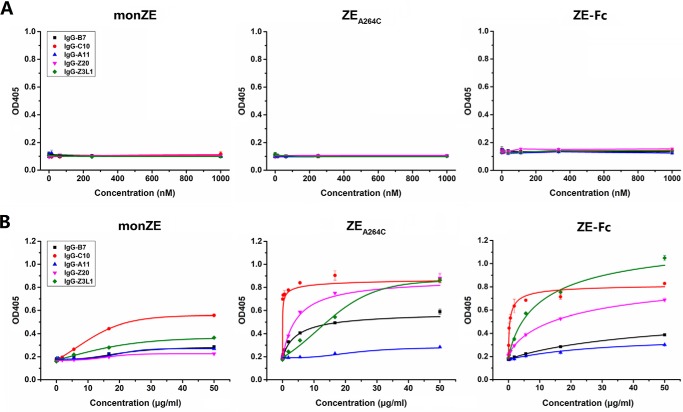
We tested the binding of IgG-Z20, IgG-Z3L1 (targeting tertiary epitopes), IgG-B7, IgG-C10, and IgG-A11 (targeting quaternary epitopes) to coated monZE, ZEA264C, and ZE-Fc. However, no binding signal was observed because the conformational epitopes might be affected (e.g. hidden or destroyed) after coating on ELISA plate (Fig. 6A). So we performed capture ELISA by coating antibodies on the plate. In general, binding of most of these antibodies (e.g. IgG-B7 and IgG-C10) to ZEA264C and ZE-Fc were stronger than that to monZE (Fig. 6B). The IgG-Z3L1 and IgG-Z20 targeting tertiary epitope recognized monZE very weakly, whereas they could strongly bind to ZEA264C and ZE-Fc. It indicates recognition of both quaternary and tertiary epitopes is highly dependent on the formation of E dimer. We also noticed that IgG-A11 does not show strong binding to monZE, ZEA264C, and ZE-Fc. In previous study, the formation of complex of E protein and Fab format IgG-A11 in vitro was also difficult (25). Probably this antibody somehow prefers to recognize epitope relying on membrane-associated E protein. According to the reports, these antibodies, isolated from convalescent patients, could neutralize ZIKV with very high potency. So, they should be able to bind to the E protein on viral surface very well. However, when monomeric E protein was used as antigen for testing, the binding was very low, indicating monZE could not present tertiary and quaternary epitopes sufficiently (27). Hence, it might be very difficult to use monomeric E protein to select antibodies that could recognize the high-order epitopes. Based on these results, it could be highly desired that use of dimeric E protein for antibodies selection would result in much higher opportunity in isolation of antibodies targeting tertiary or quaternary epitope compared with use of monomeric E protein.
Figure 6.
Recognition of tertiary/quaternary epitopes on E proteins. A, indirect ELISA. The monZE, ZEA264C, and ZE-Fc were coated on plates and serially diluted positive antibodies IgG-B7, IgG-C10, IgG-A11, IgG-Z20, and IgG-Z23L1 were added respectively. Then HRP-conjugated anti-human IgG Fc antibody was used as second antibody. B, direct sandwich ELISA. Positive antibodies were coated on plates and serially diluted monZE, ZEA264C, and ZE-Fc were added respectively. Color reaction was developed by HRP-conjugated Strep-Tactin. Error bar, the representative data are shown as mean ± S.D. from two independent experiments.
Recombinant E proteins can bind to sensitive cell lines
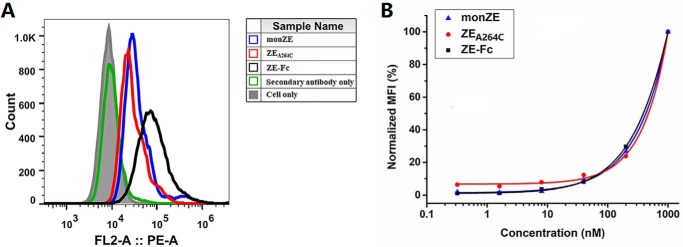
The E protein on the surface of ZIKV is essential for virus to bind and enter the host cell. Therefore, the binding ability of recombinant E proteins to sensitive cell lines is an important parameter to verify the conformation. As shown in Fig. 7A, monZE, ZEA264C, and ZE-Fc were able to bind to sensitive cell lines (e.g. bind to the cellular receptor). Although it seemed that the relative host-cell binding of ZEA264C and ZE-Fc was comparable with that of monZE (Fig. 7B), further evidence is necessary to know how the virus interacts with the host through E proteins.
Figure 7.
Flow cytometry of monZE, ZEA264C, and ZE-Fc to sensitive cells. A, Vero cells were incubated with 40 nm biotin-labeled monZE, ZEA264C, and ZE-Fc. The obvious fluorescence intensity shift was observed. As a negative control, ZEDIII showed no binding ability to cell surface at same protein concentration. B, normalized mean fluorescence intensity at different protein concentrations from 0.32 to 1000 nm. All the three proteins bound to Vero cells in a dose-dependent manner.
Comparison of immunogenicity of recombinant E proteins in mice
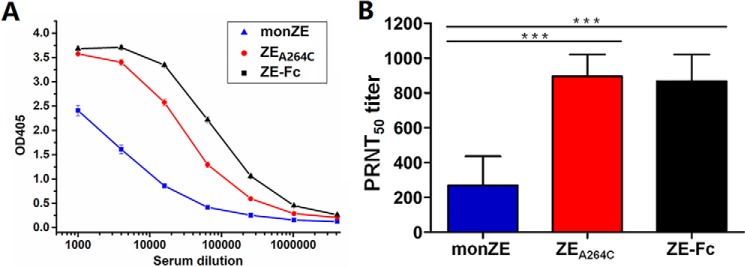
To retain the natural conformation of antigens, water-soluble adjuvant was used here. As shown in Fig. 8A, all three recombinant E proteins were able to elicit high antibody responses in mice. To be specific, the antibody titer of ZEA264C-immunized or ZE-Fc–immunized mice was 4-fold higher than that of monZE-immunized mice. The data indicate that mice immunized with dimeric E protein (ZEA264C or ZE-Fc) could produce higher antibody titer than that of monomeric E in vivo. The results of immunization in mice further disclose that dimerization of E protein could increase the immunogenicity of E protein.
Figure 8.
The antigen-specific antibody responses and neutralizing titers of immunized mice sera. A, antibody titer of different antisera. The indicated mice sera were serially diluted and then analyzed by ELISA. The different recombinant ZIKV E proteins used in immunization were used as the capture antigens. The antibody titers were defined as the last dilution showing positive readings (≥0.1 OD unit than that of the pre-immune serum). B, the neutralizing titer of antisera. The ZIKV/SZ-WIV01 strain was used to test the neutralizing potency of different antisera and the data were present as PRNT50 titer. The results were analyzed by Student's 2-tailed t test. Asterisks represent significant differences between groups:***, p <0.001. Error bar, the representative data are shown as mean ± S.D from two independent experiments.
Neutralization potency of antisera
The plaque reduction assay was employed to determine PRNT50 values of pooled sera collected from each group. As shown in Fig. 8B, all of three group antisera were able to neutralize the ZIKV/SZ-WIV01 strain, but the potency was different. The PRNT50 titers of anti-ZEA264C sera and anti-ZE-Fc sera are 896 and 867, respectively, whereas the PRNT50 value of antisera from monZE-immunized group is 269, which shows a relative low neutralizing potency.
Discussion
To control ZIKV infection, mAbs and vaccines are very promising for treatment and prophylaxis. However, how to design an ideal immunogen for selection of mAbs with high potency and development of vaccine that could induce effective humoral immune responses (e.g. generation of powerful neutralizing antibodies in vivo) is still a challenging problem (8, 10, 37, 38).
As mentioned in the introduction, mAbs targeting FLE normally cause serious ADE and their neutralizing activity is relatively modest. The mAbs that bind to DIII have reduced ADE but their neutralizing spectra are narrow. For now, it has been shown that mAbs recognizing quaternary conformation exhibit the most potent crossreactive neutralization and no obvious ADE. However, most of mAbs targeting quaternary structure E protein were isolated from convalescent patients, which is source limited. Meanwhile, because of the lack of native-like E protein, the antibodies selected from naïve phage display library or immunized animal might be modest or biased to certain epitope (20, 39). Hence, preparation of native-like E protein (E dimer) is one of the key steps to overcome these limitations. Currently, only covalent linked ZIKV E mutant (ZEA264C) has been reported to exist as dimer in solution (28). However, the detailed information on structure and function of it was still not very clear. Additionally, because native E dimer on viral surface forms noncovalently, a noncovalent dimeric E protein by fusion to antibody Fc fragment (ZE-Fc) was also designed and evaluated here.
A panel of positive antibodies was used to comprehensively characterize the recombinant protein ZEA264C and ZE-Fc. According to our results, the dimeric E could be efficiently formed under covalent or noncovalent condition and both of them could be recognized well by most of tested mAbs targeting tertiary or quaternary structure epitopes, which showed that major neutralizing epitopes including tertiary/quaternary structure epitopes are maintained in both ZEA264C and ZE-Fc. The binding activities of each tested antibody to them are comparable in general.
Because EDIII contains important neutralizing epitopes, the domain itself is a superior ZIKV vaccine candidate that is better than monZE for immunization in mice (32). The explanation for this conclusion might be that monZE could elicit relatively more antibodies targeting FLE and further generate severe ADE, which drastically decreases the protection of such kind of immunogen. However, it might be effectively improved when using dimeric E protein as an immunogen because the FL region in E dimer could be buried more than that in E monomer. In our case, ZEA264C and ZE-Fc are recognized by EDIII-specific antibody better than monZE, whereas FLE-specific antibody could bind to monZE more strongly than ZEA264C and ZE-Fc.
To further illustrate the advantages of dimeric E (ZEA264C and ZE-Fc) in vivo, immunization assay was performed in mice, and sera were obtained from different groups to assess the production of antigen-specific antibody and neutralizing potency of these antisera. First, we found ZEA264C and ZE-Fc could cause higher humoral immunity responses than monZE in vivo. Second, the antisera from ZEA264C- and ZE-Fc–immunized mice show higher neutralizing activities than that of monZE in vitro. Although we do not isolate neutralizing monoclonal antibodies from immunized mice, we could rationally speculate that stronger neutralizing potency probably derives from elicitation of tertiary/quaternary-specific antibodies, more EDIII-specific antibodies and fewer FLE-specific antibodies after immunization by ZEA264C and ZE-Fc compared with monZE.
Combined with all the results, it reasonably shows evidence that dimeric E might be a better candidate for immunogen than monomeric E. In addition, fusion with Fc fragment could bring extra benefits. It is able to enlarge molecular size and prolong the half-life of wtZE and bring other benefits in vivo after immunization, which could effectively enhance the immunogenicity and increase the reactivity of immune system (40, 41).
In conclusion, two candidate immunogens, ZEA264C and ZE-Fc, could be useful for development of therapeutic mAbs and design of vaccines against ZIKV infection. Both strategies for dimerization could be expanded to study on antiviral reagents against other flaviviruses.
Experimental procedures
Cells, virus, and antibodies
Vero cells (catalog no. CCL-81, ATCC) were grown in DMEM (Gibco) at 37 °C with 5% of CO2. Drosophila S2 cells were purchased from Invitrogen (Thermo Fisher Scientific) and cultured in Schneider's Drosophila medium (Gibco) at 28 °C. Both media were supplemented with 10% FBS (Gibco), 100 units/ml of penicillin, and 100 μg/ml streptomycin (Gibco).
The Asian lineage ZIKV strain SZ-WIV01 (GenBank accession no.: KU963796) (42) was obtained from the Microorganisms & Viruses Culture Collection Center, Wuhan Institute of Virology, Chinese Academy of Sciences. The virus was amplified in Vero cells, and titers were determined by plaque assay as described previously (42). The sequence of its E protein is identical to that of the Asian strain ZIKV H/PF/2013 (GenBank accession no.: KJ776791) which has been widely used in related studies.
The sequences of positive antibodies used here were synthesized according to the published references (Table S1). ZV-67 (anti-EDIII epitope) (20) and 2A10G6 (anti-FL epitope) (23) were in the form of single-chain variable fragment (denoted as scFv-ZV-67 and scFv-2A10G6), whereas EDE2-B7, EDE1-C10, and EDE2-A11 (25, 43) were constructed as human IgG antibodies as well as Z3L1 and Z20 (anti-EDE) (denoted as IgG-B7, IgG-C10, IgG-A11, IgG-Z3L1, and IgG-Z20) (27). The scFv-ZV-67 and scFv-2A10G6 were expressed in the Escherichia coli BL21 (DE3) cells and purified with nickel-nitrilotriacetic acid resin (Qiagen). The antibodies in human IgG form were expressed in the 293 F cells and purified with Protein A (GE Healthcare). Purified proteins were concentrated by 3-kDa (for scFv) or 30-kDa (for IgG) cut-off membrane (EMD Millipore) and concentration was measured by NanoPhotometer N60 (Implen) with corresponding extinction coefficient. All the proteins were stored at −80 °C for further analysis.
Production and purification of monZE, ZEA264C and Fc-fusion proteins
The gene encoding the E protein of ZIKV (the Asian strain H/PF/2013, GenBank accession no.: KJ776791) and an enterokinase (EK) cleavage site followed by the Twin-Strep-tag were codon-optimized and synthesized (GENEWIZ) for Drosophila S2 expression system, yielding plasmid pUC57-ZIKV-E. For producing wtZE (1–408) (25), the gene of these fragments was amplified from pUC57-ZIKV-E and cloned into the secreted expression vector pMT/BiP/V5-His A (Invitrogen), which was named as pMT-ZE-EK-Twinstrep. The single Cys mutation EA264C was introduced by SOE (gene splicing by overlap extension) PCR with primers containing mutant sequence and the construct was named as pMT-ZEA264C-EK-Twinstrep and verified by DNA sequencing. The Fc-fusion proteins were constructed by fusing wtZE or ZEA264C to Fc fragment of human IgG1 with a flexible linker (mutate all Cys residues to Ser residues), which of them were named ZE-Fc and ZEA264C-Fc, respectively.
Stable expression S2 cell lines were created as described in Drosophila Expression System User Guide (Invitrogen). Briefly, Drosophila S2 cells were co-transfected with an expression vector (pMT-ZE-EK-Twinstrep, pMT-ZEA264C-EK-Twinstrep, pMT-ZE-Fc-EK-Twinstrep, and pMT-ZEA264C-Fc-EK-Twinstrep) and the selection vector pCoBlast (Invitrogen). Blasticidin (Invitrogen) was added into Schneider's Drosophila medium to select the stable cell lines. After 2-week culture in selective medium, stable S2 cell lines expressing wtZE, ZEA264C, ZE-Fc, and ZEA264C-Fc were obtained. Then the stable cell lines were adapted into protein-free Insect-XPRESSTM Medium (Lonza) and CuSO4 was added at a final concentration of 500 μm to induce the protein expression. The supernatants were collected 7–10 days after induction and concentrated by Vivaflow 200 (Sartorius) and purified via affinity chromatography with Strep-Tactin columns (IBA) according to the manufacturer's instructions. The elution of protein was loaded into Superdex 200 10/300 GL column (GE Healthcare) equilibrated with PBS (pH 7.4) for second-step purification. Purified proteins were concentrated by 10-kDa (wtZE) or 30-kDa (ZEA264C, ZE-Fc, and ZEA264C-Fc) cut-off membrane (EMD Millipore) and concentration was measured by NanoPhotometer 60 (Implen) with corresponding extinction coefficient.
Size exclusion chromatography (SEC)
To evaluate the purity and stability of monZE, ZEA264C, and ZE-Fc proteins in solution, purified proteins were loaded into Superdex 200 10/300 GL column again (GE Healthcare) equilibrated with PBS (pH 7.4). The UV absorbance at 280 nm was monitored.
MALDI-TOF MS analysis
MALDI-MS was acquired using 5800 MALDI-TOF/TOF (Applied Biosystems/MDS Sciex) equipped with an Nd:YAG laser with 355 nm wavelength of <500 picosecond pulse and 200 Hz repetition rate. The spectrometer was operated in positive mode and the spectra were accumulated by 1200 laser shots. The MS data were further processed by using Data Explorer 4.0 (Applied Biosystems/MDS Sciex). The 50% acetonitrile/water was used to dissolve the proteins, each 1 μl of dissolved protein samples was mixed with 1 μl freshly made sinapic acid (SA) solutions (10 mg/ml in 70% MeOH). Mixtures were then loaded onto the MALDI plate and the program was performed.
SDS-PAGE
Purified proteins were analyzed by SDS-PAGE. MonZE and ZEA264C protein samples were mixed with reducing-loading buffer or nonreducing-loading buffer (Sangon Biotech). For Fc-fusion proteins, they were mixed with reducing-loading buffer and treated with boiling or nonboiling condition. Then all these samples were loaded on 10% SDS-PAGE gel. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R-250.
Circular dichroism
The secondary structures of purified proteins were determined by CD. The proteins were dissolved in PBS at the final concentration of 0.3 mg/ml, and the CD spectra were recorded from 195 to 260 nm on an Applied Photophysics Chirascan-SF.3 spectrophotometer (Applied Photophysics Ltd.) at 25 °C in a 0.1-cm pathlength cuvette (Applied Photophysics Ltd). Thermal stability was measured by recording the CD signal when temperature increased from 25 to 70 °C (monZE and ZEA264C) or from 25 to 94 °C (ZE-Fc) with a ramp rate of 0.5 °C/min at 216 nm. The CD data were shown as mean residue ellipticity.
Indirect ELISA
Purified monZE, ZEA264C, and ZE-Fc proteins were used as coating antigens, and BSA was negative control antigen. ELISA plates (Corning) were coated with 50 μl of 4 μg/ml protein overnight in PBS at 4 °C and blocked with 100 μl per well of 3% skim milk (Bio-Rad) in PBS at 37 °C for 1 h. The plates were washed with PBS containing 0.05% Tween 20 (PBST), then 3-fold serial diluted positive antibodies were added and incubated at 37 °C for 1.5 h. Plates were washed for five times with PBST and 50 μl of 1:3000 HRP-conjugated anti-His antibody (Proteintech) for scFv form or 1:5000 HRP-conjugated anti-Human IgG (Fc specific) antibody (Sigma-Aldrich) for IgG form in PBS adding 1% BSA (Sangon Biotech) per well before incubation at 37 °C for 1h. After washing with PBST, the binding was measured by the addition of diammonium 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonate) (ABTS) substrate (Invitrogen) and signal was measured at 405 nm.
Direct Sandwich ELISA
The positive antibodies were coated on the 96-well ELISA plates as capture antibodies. The methods of coating and blocking were carried out as described above. Then 3-fold serial diluted antigens (monZE, ZEA264C, and ZE-Fc) were added and incubated at 37 °C for 1.5 h. After washing, 50 μl of 1:10000 HRP-conjugated Strep-Tactin (IBA) was added and incubated at 37 °C for 1 h. The binding activity was observed with ABTS substrate and measured at 405 nm.
Cytofluorometry
Vero cells were washed and resuspended in PBS containing 1% BSA and biotin-labeled 5-fold serial diluted monZE, ZEA264C, and ZE-Fc proteins for 1 h at 4 °C. After washing, Phycoerythrin (PE)-conjugated streptavidin (Invitrogen) was added and incubated for another 1 h at 4 °C. Further analysis was carried out with a FACSCalibur (BD Biosciences). The data were processed by the software of FlowJo X.
Mice, immunization, and ethics statements
All mouse experiments were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals in China, and the protocols were approved by the ethics committee of the Wuhan Institute of Virology, Chinese Academy of Science (permit number WIVA34201702).
Female BALB/c mice (6–8 weeks) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were housed in a specific pathogen-free animal laboratory under standard conditions.
Prior to immunization, purified monZE, ZEA264C, and ZE-Fc proteins were mixed well with QuickAntibody-Mouse5W adjuvant (Biodragon Immunotech). Each injection dose contained 10 μg protein and 50 μl adjuvant in a volume of 100 μl. In addition, PBS was mixed with same volume adjuvant as the negative control. Mice were divided into four groups (n = 6): monZE, ZEA264C, ZE-Fc, and PBS. The mixture was injected into a quadriceps muscle of each mouse. After 3 weeks, a boost immunization was performed with the same dose and method. Blood samples were collected from the mice at 2 weeks after the final immunization and sera were isolated for further evaluation.
Antibody titer in sera
The antigen-specific serum antibody titers were measured by ELISA. Briefly, ELISA plate was coated with monZE, ZEA264C, and ZE-Fc respectively overnight at 4 °C. Next day, the wells were blocked and then 4-fold serial dilutions (starting at 1:1000) of pooled mouse serum were added for 1.5 h at 37 °C, followed by incubation with 50 μl/well of HRP-conjugated goat anti-mouse IgG antibody (1:50,000 diluted in 1% BSA, Abcam) for 1 h at 37 °C. After color development, the absorbance was measured at 405 nm and end point titer was reported as the reciprocal of the highest serum dilution that had an absorbance ≥0.1 optical density (OD) unit above that of the pre-immune samples.
Virus neutralization assay
The neutralization activities of immunized mouse sera were determined by performing plaque reduction assay. Briefly, pooled mouse sera were 3-fold serially diluted, starting at a dilution of 1:50, and then diluted serum samples were mixed with 100 plaque forming units ZIKV solution, followed by incubation at 37 °C for 1 h. The serum/virus mixtures were added onto pre-seeded Vero cell monolayers in 24-well plates and incubated at 37 °C for 1.5 h. Then, the medium was replaced with fresh DMEM containing 2% FBS and 2% methylcellulose. The plates were then transferred to 37 °C in 5% CO2 incubator. At 80 h post infection, plaques were visualized by fixation with 4% paraformaldehyde and staining with 0.1% crystal violet. For a given serum sample, the percent reduction of plaques was calculated by comparing the plaque number obtained to that of the virus only. The 50% plaque reduction neutralization titers (PRNT50) were determined by a four-parameter logistic regression.
Author contributions
C. Y. and R. G. formal analysis; C. Y., F. Z., X. G., S. Z., X. L., S. L., N. L., and C. D. methodology; C. Y. and R. G. writing-original draft; C. Y., B. Z., and R. G. writing-review and editing; R. G. supervision; R. G. funding acquisition; R. G. project administration.
Supplementary Material
Acknowledgments
We are thankful to the Core Facility and Technical Support, Wuhan Institute of Virology, Chinese Academy of Sciences; Wuhan Institute of Biotechnology; Wuhan Key Laboratory on Emerging Infectious Diseases and Biosafety; as well as Wuhan National Bio-Safety Level 4 Lab of the Chinese Academy of Sciences for the support.
This work was funded by the External Cooperation Program of Chinese Academy of Sciences Grant 153211KYSB20160001, the Key Program of Chinese Academy of Sciences Grant ZDRW-ZS-2016-4, the National Key Research and Development Program of China Grant 2016YFC1202902, and the “One-Three-Five” Strategic Programs of Wuhan Institute of Virology, Chinese Academy of Sciences Grant WIV-135-PY4. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
- ZIKV
- Zika virus
- ADE
- antibody-dependent enhancement
- E
- envelope
- EDE
- E-dimer–dependent epitope
- FL
- fusion loop
- FLE
- FL epitope
- M.M.
- molecular mass
- PBST
- PBS containing 0.05% Tween 20
- PRNT50
- 50% plaque reduction neutralization titer
- SEC
- size exclusion chromatography.
References
- 1. Lazear H. M., and Diamond M. S. (2016) Zika virus: New clinical syndromes and its emergence in the western hemisphere. J. Virol. 90, 4864–4875 10.1128/JVI.00252-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medin C. L., and Rothman A. L. (2017) Zika virus: The agent and its biology, with relevance to pathology. Arch. Pathol. Lab. Med. 141, 33–42 10.5858/arpa.2016-0409-RA [DOI] [PubMed] [Google Scholar]
- 3. Brasil P., Sequeira P. C., Freitas A. D., Zogbi H. E., Calvet G. A., de Souza R. V., Siqueira A. M., de Mendonca M. C., Nogueira R. M., de Filippis A. M., and Solomon T. (2016) Guillain-Barré syndrome associated with Zika virus infection. Lancet 387, 1482 10.1016/S0140-6736(16)30058-7 [DOI] [PubMed] [Google Scholar]
- 4. Bautista L. E., and Sethi A. K. (2016) Association between Guillain-Barré syndrome and Zika virus infection. Lancet 387, 2599–2600 10.1016/S0140-6736(16)30844-3 [DOI] [PubMed] [Google Scholar]
- 5. de Oliveira W. K., de França G. V. A., Carmo E. H., Duncan B. B., de Souza Kuchenbecker R., and Schmidt M. I. (2017) Infection-related microcephaly after the 2015 and 2016 Zika virus outbreaks in Brazil: A surveillance-based analysis. Lancet 390, 861–870 10.1016/S0140-6736(17)31368-5 [DOI] [PubMed] [Google Scholar]
- 6. Lucey D., Cummins H., and Sholts S. (2017) Congenital Zika syndrome in 2017. JAMA 317, 1368–1369 10.1001/jama.2017.1553 [DOI] [PubMed] [Google Scholar]
- 7. Meneses J. D. A., Ishigami A. C., de Mello L. M., de Albuquerque L. L., de Brito C. A. A., Cordeiro M. T., and Pena L. J. (2017) Lessons learned at the epicenter of Brazil's congenital Zika epidemic: Evidence from 87 confirmed cases. Clin. Infect. Dis. 64, 1302–1308 10.1093/cid/cix166 [DOI] [PubMed] [Google Scholar]
- 8. Lin H. H., Yip B. S., Huang L. M., and Wu S. C. (2018) Zika virus structural biology and progress in vaccine development. Biotechnol. Adv. 36, 47–53 10.1016/j.biotechadv.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 9. Munjal A., Khandia R., Dhama K., Sachan S., Karthik K., Tiwari R., Malik Y. S., Kumar D., Singh R. K., Iqbal H. M. N., and Joshi S. K. (2017) Advances in developing therapies to combat Zika virus: Current knowledge and future perspectives. Front. Microbiol. 8, 1469 10.3389/fmicb.2017.01469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie X., Zou J., Shan C., and Shi P. Y. (2017) Small molecules and antibodies for Zika therapy. J. Infect. Dis. 216, S945–S950 10.1093/infdis/jix406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sirohi D., and Kuhn R. J. (2017) Zika virus structure, maturation, and receptors. J. Infect. Dis. 216, S935–S944 10.1093/infdis/jix515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasan S. S., Sevvana M., Kuhn R. J., and Rossmann M. G. (2018) Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 25, 13–20 10.1038/s41594-017-0010-8 [DOI] [PubMed] [Google Scholar]
- 13. Sirohi D., Chen Z., Sun L., Klose T., Pierson T. C., Rossmann M. G., and Kuhn R. J. (2016) The 3.8 A resolution cryo-EM structure of Zika virus. Science 352, 467–470 10.1126/science.aaf5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stiasny K., and Heinz F. X. (2006) Flavivirus membrane fusion. J. Gen. Virol. 87, 2755–2766 10.1099/vir.0.82210-0 [DOI] [PubMed] [Google Scholar]
- 15. Smit J. M., Moesker B., Rodenhuis-Zybert I., and Wilschut J. (2011) Flavivirus cell entry and membrane fusion. Viruses 3, 160–171 10.3390/v3020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chao L. H., Klein D. E., Schmidt A. G., Peña J. M., and Harrison S. C. (2014) Sequential conformational rearrangements in flavivirus membrane fusion. eLife 3, e04389 10.7554/eLife.04389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kostyuchenko V. A., Lim E. X., Zhang S., Fibriansah G., Ng T. S., Ooi J. S., Shi J., and Lok S. M. (2016) Structure of the thermally stable Zika virus. Nature 533, 425–428 10.1038/nature17994 [DOI] [PubMed] [Google Scholar]
- 18. Sevvana M., Long F., Miller A. S., Klose T., Buda G., Sun L., Kuhn R. J., and Rossmann M. G. (2018) Refinement and analysis of the mature Zika virus cryo-EM structure at 3.1 A resolution. Structure 26, 1169–1177.e3 10.1016/j.str.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pierson T. C., and Kielian M. (2013) Flaviviruses: Braking the entering. Curr. Opin. Virol. 3, 3–12 10.1016/j.coviro.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao H., Fernandez E., Dowd K. A., Speer S. D., Platt D. J., Gorman M. J., Govero J., Nelson C. A., Pierson T. C., Diamond M. S., and Fremont D. H. (2016) Structural basis of Zika virus-specific antibody protection. Cell 166, 1016–1027 10.1016/j.cell.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stettler K., Beltramello M., Espinosa D. A., Graham V., Cassotta A., Bianchi S., Vanzetta F., Minola A., Jaconi S., Mele F., Foglierini M., Pedotti M., Simonelli L., Dowall S., Atkinson B., et al. (2016) Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826 10.1126/science.aaf8505 [DOI] [PubMed] [Google Scholar]
- 22. Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., Jumnainsong A., Edwards C., Quyen N. T. H., Duangchinda T., Grimes J. M., Tsai W. Y., Lai C. Y., Wang W. K., Malasit P., et al. (2015) A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 10.1038/ni.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dai L., Song J., Lu X., Deng Y. Q., Musyoki A. M., Cheng H., Zhang Y., Yuan Y., Song H., Haywood J., Xiao H., Yan J., Shi Y., Qin C. F., Qi J., and Gao G. F. (2016) Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 10.1016/j.chom.2016.04.013 [DOI] [PubMed] [Google Scholar]
- 24. Swanstrom J. A., Plante J. A., Plante K. S., Young E. F., McGowan E., Gallichotte E. N., Widman D. G., Heise M. T., de Silva A. M., and Baric R. S. (2016) Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. MBio 7, e01123–16 10.1128/mBio.01123-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barba-Spaeth G., Dejnirattisai W., Rouvinski A., Vaney M. C., Medits I., Sharma A., Simon-Lorière E., Sakuntabhai A., Cao-Lormeau V. M., Haouz A., England P., Stiasny K., Mongkolsapaya J., Heinz F. X., Screaton G. R., and Rey F. A. (2016) Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53 10.1038/nature18938 [DOI] [PubMed] [Google Scholar]
- 26. Hasan S. S., Miller A., Sapparapu G., Fernandez E., Klose T., Long F., Fokine A., Porta J. C., Jiang W., Diamond M. S., Crowe J. E. Jr., Kuhn R. J., and Rossmann M. G. (2017) A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun. 8, 14722 10.1038/ncomms14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q., Yang H., Liu X., Dai L., Ma T., Qi J., Wong G., Peng R., Liu S., Li J., Li S., Song J., Liu J., He J., Yuan H., et al. (2016) Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci. Transl. Med. 8, 369ra179 10.1126/scitranslmed.aai8336 [DOI] [PubMed] [Google Scholar]
- 28. Slon Campos J. L., Marchese S., Rana J., Mossenta M., Poggianella M., Bestagno M., and Burrone O. R. (2017) Temperature-dependent folding allows stable dimerization of secretory and virus-associated E proteins of dengue and Zika viruses in mammalian cells. Sci. Rep. 7, 966 10.1038/s41598-017-01097-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Metz S. W., Gallichotte E. N., Brackbill A., Premkumar L., Miley M. J., Baric R., and de Silva A. M. (2017) In vitro assembly and stabilization of dengue and Zika virus envelope protein homo-dimers. Sci. Rep. 7, 4524 10.1038/s41598-017-04767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rouvinski A., Dejnirattisai W., Guardado-Calvo P., Vaney M. C., Sharma A., Duquerroy S., Supasa P., Wongwiwat W., Haouz A., Barba-Spaeth G., Mongkolsapaya J., Rey F. A., and Screaton G. R. (2017) Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat. Commun. 8, 15411 10.1038/ncomms15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang C., Gao X., and Gong R. (2017) Engineering of Fc fragments with optimized physicochemical properties implying improvement of clinical potentials for Fc-based therapeutics. Front. Immunol. 8, 1860 10.3389/fimmu.2017.01860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qu P., Zhang W., Li D., Zhang C., Liu Q., Zhang X., Wang X., Dai W., Xu Y., Leng Q., Zhong J., Jin X., and Huang Z. (2018) Insect cell-produced recombinant protein subunit vaccines protect against Zika virus infection. Antiviral Res. 154, 97–103 10.1016/j.antiviral.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 33. Kostyuchenko V. A., Zhang Q., Tan J. L., Ng T. S., and Lok S. M. (2013) Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87, 7700–7707 10.1128/JVI.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blazevic J., Rouha H., Bradt V., Heinz F. X., and Stiasny K. (2016) Membrane anchors of the structural flavivirus proteins and their role in virus assembly. J. Virol. 90, 6365–6378 10.1128/JVI.00447-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saphire E. O., Parren P. W., Pantophlet R., Zwick M. B., Morris G. M., Rudd P. M., Dwek R. A., Stanfield R. L., Burton D. R., and Wilson I. A. (2001) Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science 293, 1155–1159 10.1126/science.1061692 [DOI] [PubMed] [Google Scholar]
- 36. Zeng F., Yang C., Gao X., Li X., Zhang Z., and Gong R. (2018) Comprehensive elucidation of the structural and functional roles of engineered disulfide bonds in antibody Fc fragment. J. Biol. Chem. 293, 19127–19135 10.1074/jbc.RA118.005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barouch D. H., Thomas S. J., and Michael N. L. (2017) Prospects for a Zika virus vaccine. Immunity 46, 176–182 10.1016/j.immuni.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang C., Gong R., and de Val N. (2019) Development of neutralizing antibodies against Zika virus based on its envelope protein structure. Virol. Sin. 34, 168–174 10.1007/s12250-019-00093-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Y., Li S., Du L., Wang C., Zou P., Hong B., Yuan M., Ren X., Tai W., Kong Y., Zhou C., Lu L., Zhou X., Jiang S., and Ying T. (2017) Neutralization of Zika virus by germline-like human monoclonal antibodies targeting cryptic epitopes on envelope domain III. Emerg. Microbes Infect. 6, e89 10.1038/emi.2017.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L. (2018) Pharmacokinetics of monoclonal antibodies and Fc-fusion proteins. Protein Cell 9, 15–32 10.1007/s13238-017-0408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye L., Zeng R., Bai Y., Roopenian D. C., and Zhu X. (2011) Efficient mucosal vaccination mediated by the neonatal Fc receptor. Nat. Biotechnol. 29, 158–163 10.1038/nbt.1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng C., Liu S., Zhang Q., Xu M., Zhang H., Gu D., Shi L., He J., Xiao G., and Zhang B. (2016) Isolation and characterization of Zika virus imported to China using C6/36 mosquito cells. Virol. Sin. 31, 176–179 10.1007/s12250-016-3778-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M. C., Kikuti C. M., Navarro Sanchez M. E., Dejnirattisai W., Wongwiwat W., Haouz A., Girard-Blanc C., Petres S., Shepard W. E., Desprès P., Arenzana-Seisdedos F., et al. (2015) Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113 10.1038/nature14130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.