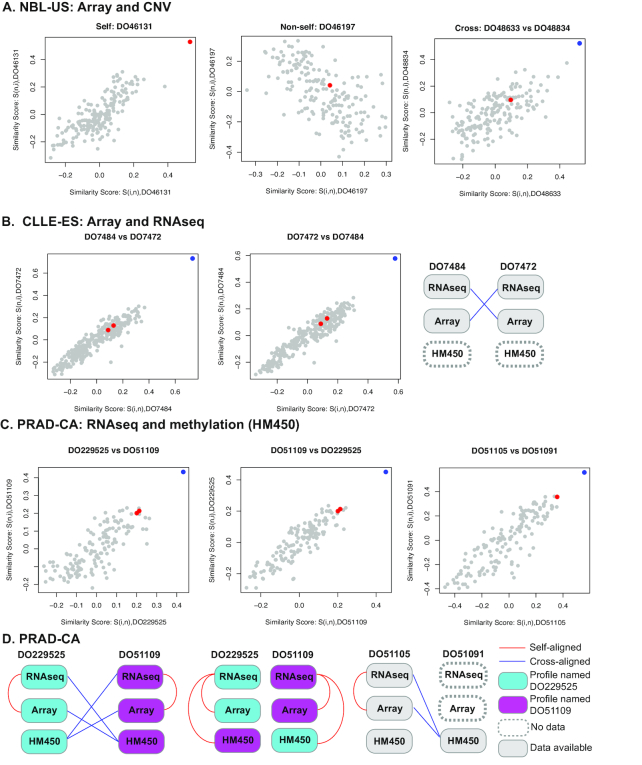
Figure 9.
Application to ICGC datasets (A) An example of self-self aligned, non–self-self aligned, and cross-aligned pairs of samples based on alignment between Array and CNV profiles in the NBL-US dataset. (B) An example of sample-labeling errors. In alignment of Array and DNA methylation profiles, DO7484 and DO7472 were cross-aligned to each other. The similarity scores of each cross-alignment are shown. The similarity score of the cross-aligned pair is shown in blue, and the similarity scores of self-self alignments are shown in red. Omics profiles of DO7484 and DO7472 were compared with each other and results were summarized into a patient-centric view. The red line indicates self-aligned, the and blue line indicates cross-aligned. (C) An example of possible sample swaps and sample-labeling errors. DO229525 and DO51109 were cross-aligned to each other in alignment of RNAseq and DNA methylation profiles as well as Array and DNA methylation profiles. Additionally, the RNAseq and Array profiles of DO51105 were cross-aligned to the DNA methylation profile of DO51091. (D) Other omics profiles of these pairs were compared with each other and results were summarized into a patient-centric view. After swapping the corresponding DNA methylation profiles, multiple-omics profiles of DO229525 and DO51109 were aligned to each other consistently.