Figure 1. Identified sites of phosphorylation in A. fumigatus CrzA and comparison of phosphopeptide abundance in wild-type and ΔcnaA backgrounds.
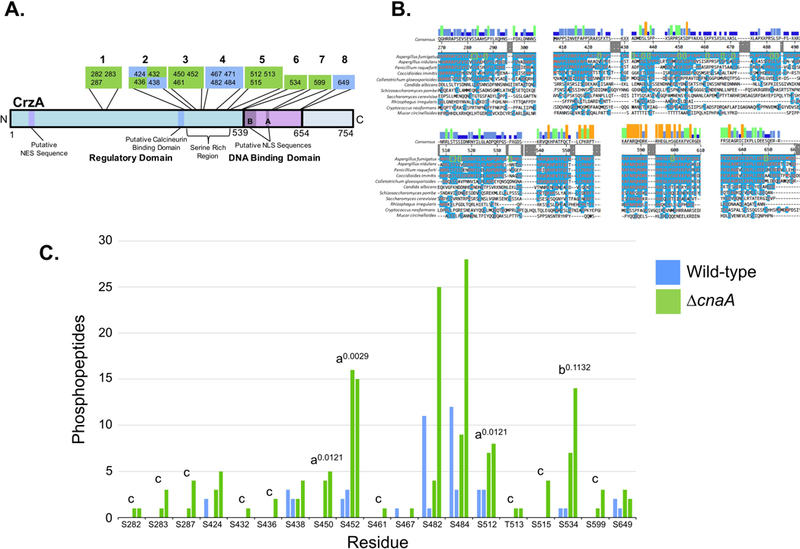
20 CrzA amino acid residues were positively identified as phosphorylated via LC-MS/MS. (A) Diagram of phosphorylation site locations in A. fumigatus CrzA. The 20 sites were grouped into 8 clusters. Sites in green boxes were phosphorylated in a calcineurin-dependent or potentially calcineurin-dependent manner while phosphorylation of sites in blue boxes was categorized as calcineurin-independent. Putative functional regions of the protein are labeled. NES = Nuclear Export Sequence, NLS = Nuclear Localization Sequence, SRR = Serine Rich Region. (B) Clustal W alignment of A. fumigatus CrzA and fungal homologues at phosphorylated regions. Blue highlighting of residues indicates identity with A. fumigatus sequence. Phosphorylated sites are indicated by green boxes. Bars above consensus sequence indicate degree of conservation (blue = less conserved, red = more conserved). (C) Number of peptides with phosphorylation at corresponding sites identified with at least 95% localization probability for each duplicate sample in the wild-type (blue) and ΔcnaA (green) genetic backgrounds. aIndicates statistically significant enrichment in ΔcnaA background compared to wild-type (student’s T-test, P < 0.05). Superscript numbers indicate specific P-values. bIndicates site with non-statistically significant enrichment in ΔcnaA background compared to wild-type still suggestive of potential calcineurin-dependent dephosphorylation. Superscript number indicates specific P-value. cIndicates sites only identified in the ΔcnaA background but for which statistically significant differences between genetic backgrounds could not be demonstrated.
