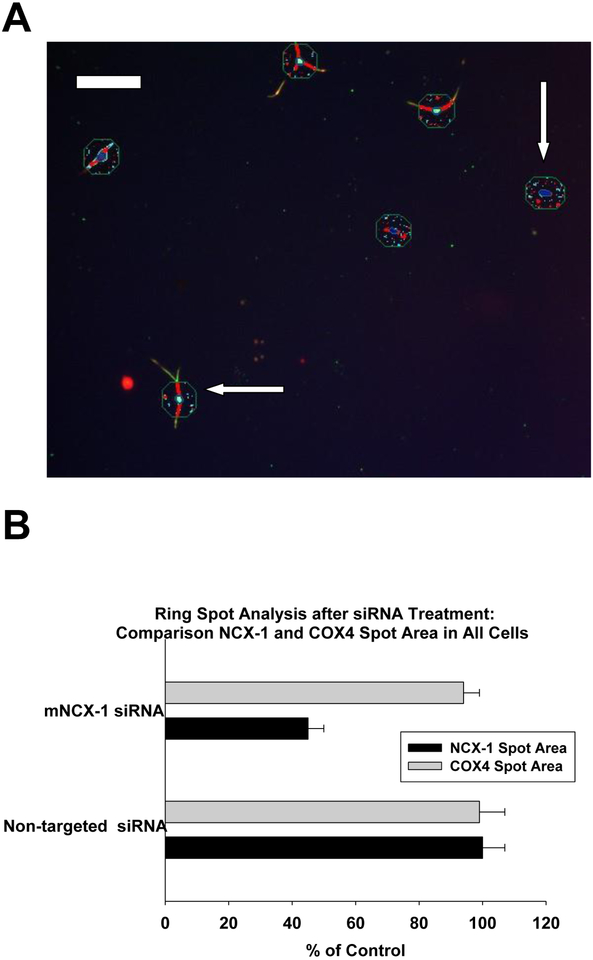
Fig. 5.
Ring spot image analysis of dorsal root ganglion cultures on day 9 after plating (part A). The punctate spot imaging is of the cytoplasmic domain and the nucleus of all cells. Mitochondria-related spots (blue) were identified with COX-4 immunofluorescence and mNCX-1 –related spots are shown in red. Both neuronal cells (horizontal arrow) and non-neuronal cells (vertical arrow) are shown for comparison. This ring spot region was initially chosen because of the predominant location of mitochondria in the cell bodies of neurons, and the non-neuronal cells were displayed for comparative purposes. The calibration bar shown in panel A represents 100 μ. In panel B, a comparison of mNCX-1 spot area (black bars) and COX-4 spot area (gray bars) are shown for all cells in cultures treated with siRNA to mNCX-1 or to non-targeted siRNA. All data are expressed as a percentage of values from control cultures. The siRNA treatment paradigm was conducted as described in figure 1. The error bars are standard errors and the bars represent the mean of 5 determinations from a representative experiment from two replicate studies.