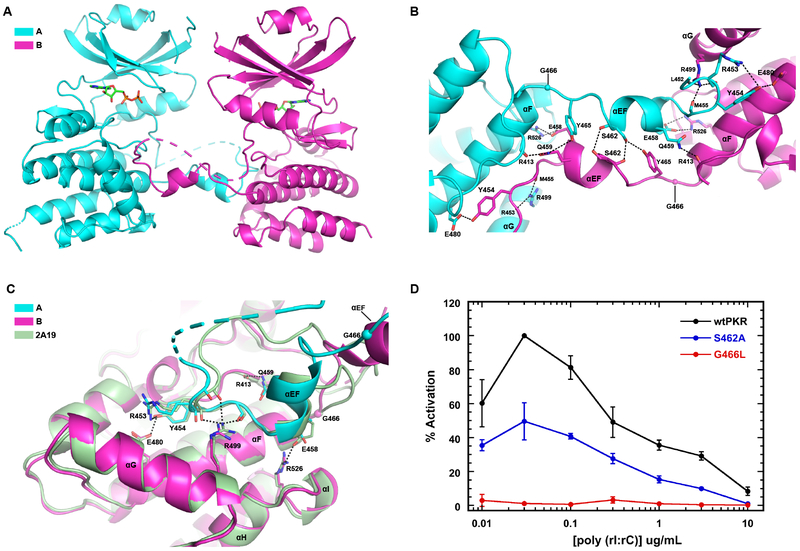
Figure 3. Analysis of the PKR kinase face-to-face dimer with exchange of activation loops.
A) Structure of the interface. The A and B chains of the AMPPNP complex of PKR kinase are depicted using the color scheme from Figure 1. The protomers are indicated in cartoon representation with the disordered regions of the activation loop and the C-terminus shown as dashes. The bound nucleotide is depicted in stick representation. B) Detailed view of the interactions stabilizing the interface. Key side chain and main chain atoms are rendered as sticks. Hydrogen bond and salt-bridge interactions are denoted by dashed lines. G466 is shown as a sphere. C) Structural alignment of a monomeric, phosphorylated PKR kinase (2A19) onto chain B forming a domain-swapped FTF dimer with chain A. The side chain and main chain atoms involved in polar interactions at the interface are rendered as sticks. D) Effect of interface mutations on PKR activation. The PKR autophosphorylation activity was assayed as a function of dsRNA concentration. The data are normalized to the maximal activation of wild-type PKR.