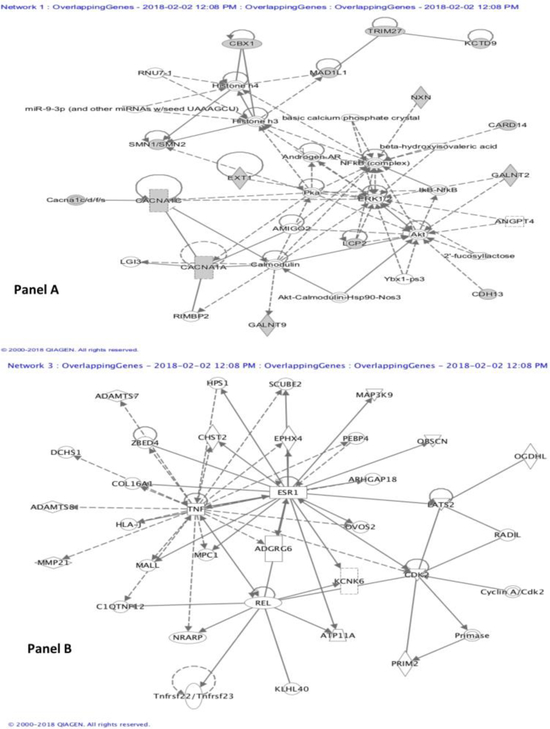
Figure 2:
Gene-gene interaction networks. Genes associated with the differentially methylated sites were uploaded to Ingenuity pathway analysis. Based on p-value cutoffs of 10−8, three networks were identified. Two of them were similar in function with several overlapping molecules. Hence, two of the different networks are presented here. The network in panel A is associated with Cell Signaling, Molecular Transport, Vitamin and Mineral Metabolism. It had a p-score of 33 and 14 focus molecules (including CACNA1A, CACNA1C, Calmodulin, ERK1/2, Histone h3, Histone h4, IkB-NfkB, NFkB (complex), miR-9–3p). The network in Panel B is associated with Neurological Disease, Organismal Injury and Abnormalities, Connective Tissue Development and Function; with a p-score of 12, and 6 focus molecules (including ESR1, KCNK6, PRIM2, TNF).