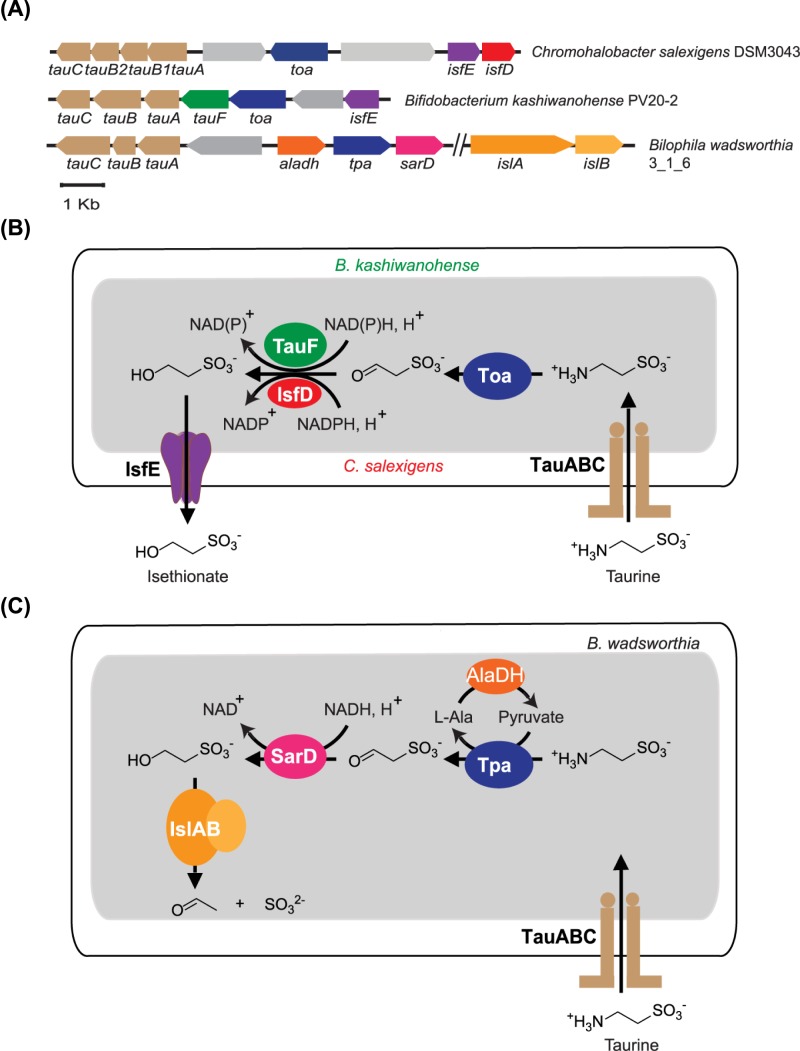
Figure 1. Gene clusters and metabolic pathways involving different sulfoacetaldehyde reductases isozymes.
(A) Gene clusters containing the sulfoacetaldehyde reductases IsfD (in Chromohalobacter salexigens), TauF (in Bifidobacterium kashiwanohense) and SarD (in Bilophila wadsworthia). (B) Schematic diagram showing two bacterial taurine nitrogen assimilation pathways relying on different sulfoacetaldehyde reductase isozymes. The pathway in C. salexigens relies on IsfD (red), while the putative pathway in B. kashiwanohense relies on TauF (green). (C) Schematic diagram showing the taurine dissimilation pathway in B. wadsworthia.