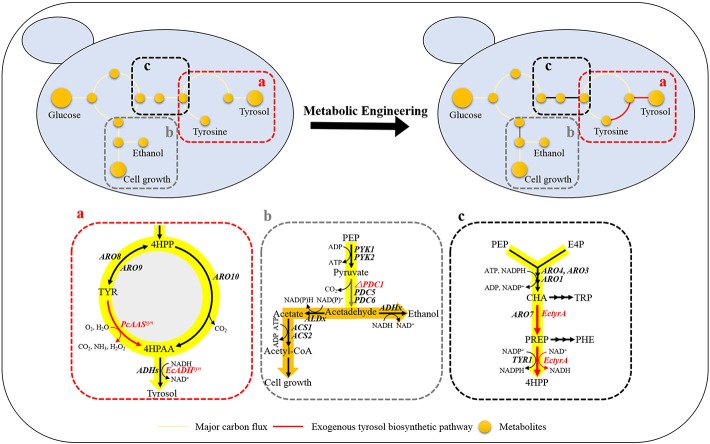
Figure 1.
Design of rational metabolic engineering to enhance tyrosol production in S. cerevisiae. Metabolites: 4HPP, 4-hydroxyphenylpyruvate; TYR, tyrosine; 4HPAA, 4-hydroxyphenylacetaldehyde; PEP, phosphoenolpyruvate; Acetyl-CoA, acetyl coenzyme A; E4P, erythrose-4-phosphate; CHA, chorismate; PREP, prephenate; TRP, tryptophan; and PHE, phenylalanine. Genes: aminotransferase gene (ARO8 and ARO9); phenylpyruvate decarboxylase gene(ARO10); pyruvate kinase gene (PYK1 and PYK2); pyruvate decarboxylase genes (PDC1, PDC5, and PDC6); chorismate mutase gene (ARO7); 3-deoxy-7-phosphoheptulonate synthase (ARO4 and ARO3); pentafunctional AROM polypeptide (ARO1); the NADP+-dependent prephenate dehydrogenase gene (TYR1); alcohol dehydrogenase genes (ADHs). The codon-optimized aromatic aldehyde synthase gene from Petroselinum crispum (PcAASsyn) and the NADH-dependent aryl-alcohol dehydrogenase gene from E. coli (EcADHsyn) were heterologously expressed in S. cerevisiae to convert tyrosine to 4HPAA (a), the native pyruvate decarboxylase gene PDC1 was knocked out for tuning down the carbon flux below the pyruvate node (b), the NAD+-dependent fused chorismate mutase/prephenate dehydrogenase gene from Escherichia coli (EctyrA) was expressed heterologously in S. cerevisiae to enhance choristmate metabolism (c).