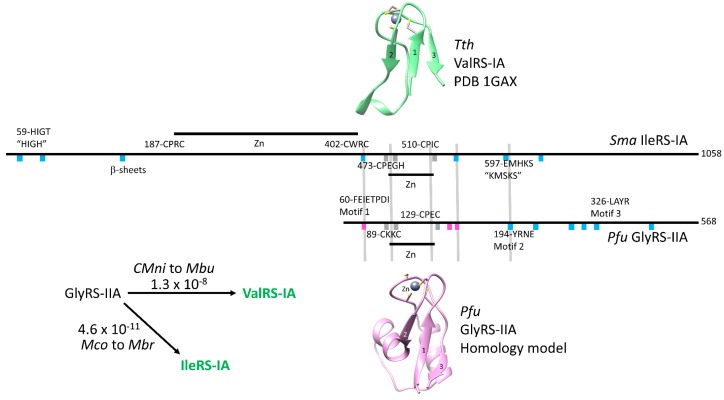
Figure 8.
A schematic sequence alignment of a class IIA aaRS and a class IA aaRS is shown [2,5]. The shared Zn fingers are shown as structures. Cyan blocks indicate active site β-sheets. Grey blocks indicate shared β-sheets that organize Zn fingers. Magenta blocks indicate β-sheets surrounding the Zn finger in Pyrococcus furiosis (Pfu) GlyRS-IIA that are organized by the Zn finger. Tth indicates Thermus thermophilus. NCBI tools were used to find close matches comparing GlyRS-IIA and IleRS-IA and GlyRS-IIA and ValRS-IA, in order to demonstrate homology of class I and class II aaRS enzymes. e-values are shown. The species compared are indicated: Candidatus Methanoperedens nitroreducens (CMni), Methanococcoides burtonii (Mbu), Methanobacterium congolense (Mco), and Methanobacterium bryantii (Mbr). Green type indicates ValRS-IA and IleRS-IA have editing active sites.