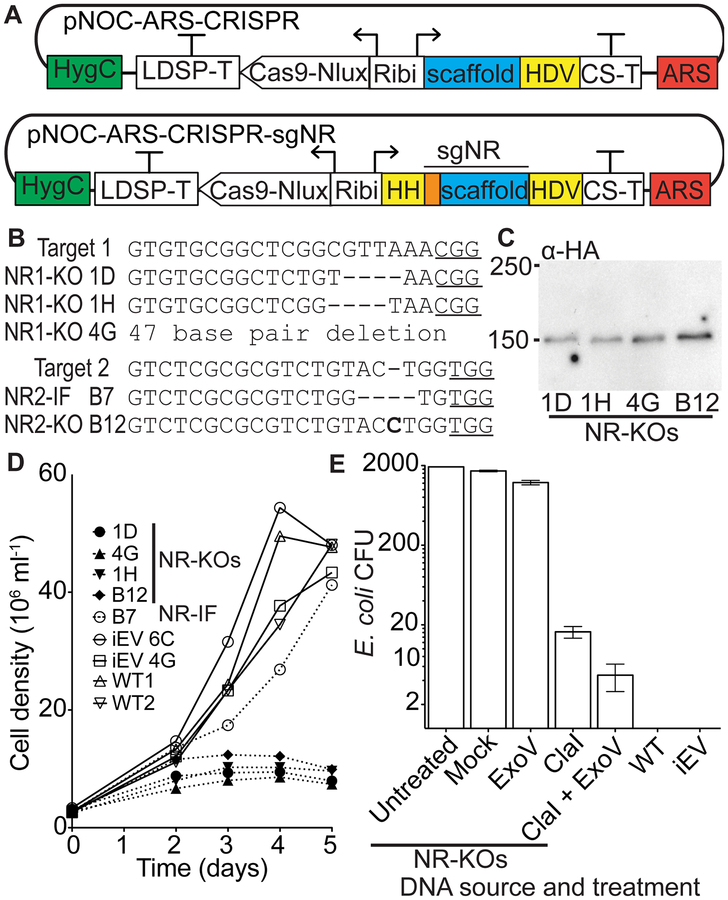
Figure 2.
Development of an episomal CRISPR system. (A) The S. cerevisiae CEN/ARS6 region (ARS, red) was included in the pNOC-ARS-CRISPR construct for episomal maintenance. Guide sequences(orange) for nitrate reductase targets, with a 5’hammerhead ribozyme (HH),were fused to the gRNA scaffold to form the NR sgRNAs (sgNR). Ribozymes are highlighted in yellow. The sgNR1 and sgNR2were added to pNOC-ARS-CRISPR to form pNOC-ARS-CRISPR-sgNR1 and pNOC-ARS-CRISPR-sgNR2, respectively. N. oceanica is transformed with circular episomal CRISPR constructs. (B) Mutations in the two target sites in the NR genomic locus (Target 1 and Target 2) of N. oceanica transformed with the respective pNOC-ARS-CRISPR-sgNR construct. Mutant lines are identified by 96-well plate location (Figure S3). Deleted nucleotides are represented with dashes and inserted nucleotides are shown in bold. Protospacer adjacent motifs (PAM sites) are underlined. (C) Immunoblot using an α-HA antibody of N. oceanicaNR1-KO and NR2-KO lines producing Cas9-Nlux-HA from the CRISPR episome. Numbers on the left indicate size markers (KDa). (D) Growth curves after transfer from NH4 to NO3 containing medium of NR-KO frame-shifted lines (1D, 1H, 4G, B12) and NR2-IF B7 in-frame line, empty vector integrated CRISPR control lines (iEV), and wildtype (WT). (E) Episome rescue by E. coli transformation using equal quantities of DNA isolated from episomal (NR-KO) lines, integrated empty-vector (iEV), and wild-type (WT) N. oceanica lines. Values are the average colonies generated ± SE (NR-KOs n = 3 independent lines, WT n = 2 biological replicates, and iEV n = 2 independent lines). Equal quantities of DNA from NR-KO lines after treatment were used for E. coli transformation, and the resulting colonies counted (n = 3 independent lines). Exonuclease V (ExoV), ClaI endonuclease (ClaI), and ClaI endonuclease with Exonuclease V (ClaI+ExoV).