Figure 1.
Proteomic Analysis of Upregulated Proteins Highlights BH4/NO Modulation of Inflammation, Mitochondrial Function, and Metabolism
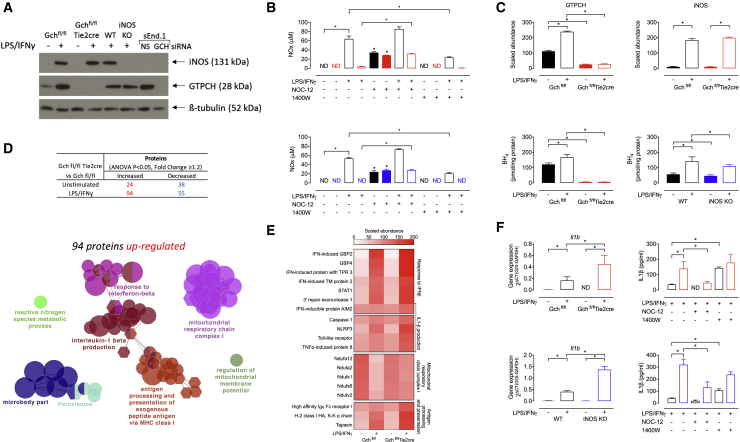
(A) Western blot analysis of iNOS and GTPCH protein levels in Gchfl/flTie2cre and iNOS KO BMDMs stimulated with LPS and IFNγ for 16 h. β-Tubulin was used as a loading control. sEnd.1 murine endothelial cells transfected with non-specific (NS) and GCH targeted siRNA were used as positive and negative controls for GTPCH (n = 3).
(B) NOx (nitrite+nitrate) accumulation in the media measured using an NO analyzer (n = 5–6) (Black bars, Gchfl/fl or WT; Red bars, Gchfl/flTie2cre; Blue bars, iNOS KO).
(C) The abundance of GTPCH and iNOS proteins were determined by mass spectrometry (n = 4) and intracellular BH4 quantified using HPLC (n = 5–6).
(D) Number of proteins significantly (ANOVA, p < 9x10−6) changed in abundance as determined by mass spectrometric analysis (n = 4) and GO term enrichment analysis of significantly upregulated proteins in Gchfl/flTie2cre BMDMs stimulated with LPS/IFNγ. The most significant term in each cluster is annotated.
(E) Heatmap showing the scaled abundance of proteins identified in the mostly significantly enriched GO terms from the analysis of upregulated proteins.
(F) Il1b gene expression in Gchfl/flTie2cre and iNOS KO cells measured using qRT-PCR (n = 5) and levels of IL-1β in supernatants from Gchfl/flTie2cre and iNOS KO cells measured by ELISA (n = 5).
Data are mean + SEM; p values calculated using 2-way ANOVA with Tukey’s post-test (∗p < 0.05).