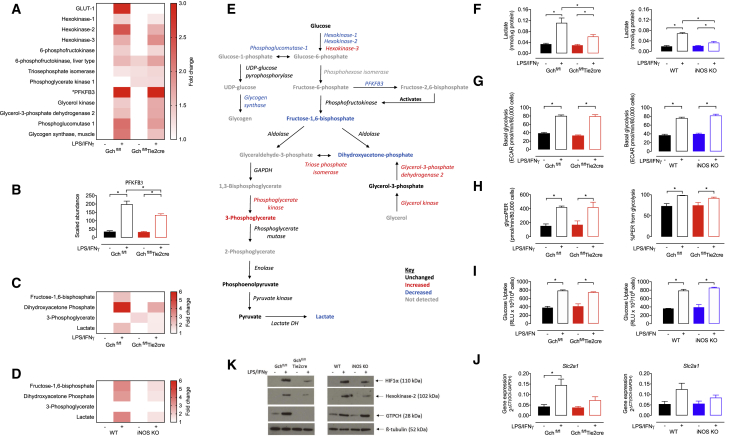
Figure 4.
Glycolytic Rate Is Unaffected by Loss of NO Signaling Despite Changes in the Levels of Glycolytic Proteins and Metabolites
(A) Heatmap showing fold changes in abundance of enzymes significantly changed relative to Gchfl/fl unstimulated cells (n = 4, p < 0.05, ∗PFKB3 = fold change > 3).
(B) Abundance of PFKFB3 protein (n = 4).
(C and D) (C) Fold change heatmaps of significantly changed (p > 0.05) metabolites relative to unstimulated Gchfl/fl or (D) unstimulated WT cells (n = 6).
(E) Glucose metabolism pathway representing changes (>20%) in abundance of enzymes and metabolites in LPS/IFNγ stimulated Gchfl/flTie2cre versus Gchfl/fl macrophages.
(F) Lactate accumulation measured in medium supernatants from macrophages following overnight stimulation (n = 4).
(G) Basal glycolysis determined using 2-deoxyglucose inhibitable ECAR measured using XFe96 Seahorse bioanalyzer in Gchfl/flTie2cre and iNOS KO cells (n = 6).
(H) Basal glycolytic rate determined in Gchfl/flTie2cre cells by calculating glycolytic proton efflux rate (glycoPER) (n = 3).
(H) The contribution of glycolysis to acidification of media in Gchfl/flTie2cre cells (n = 3).
(I) Measurement of glucose uptake in Gchfl/flTie2cre and iNOS KO cells (n = 4).
(J) qRT-PCR mRNA measurement of Slc2a1 gene encoding GLUT-1 protein in Gchfl/flTie2cre and iNOS KO cells (n = 5).
(K) Western blot analysis of HIF1α and hexokinase 2 proteins in Gchfl/flTie2cre and iNOS KO cells (n = 4).
Data are mean + SEM; p values calculated using 2-way ANOVA with Tukey’s post-test (∗p < 0.05).