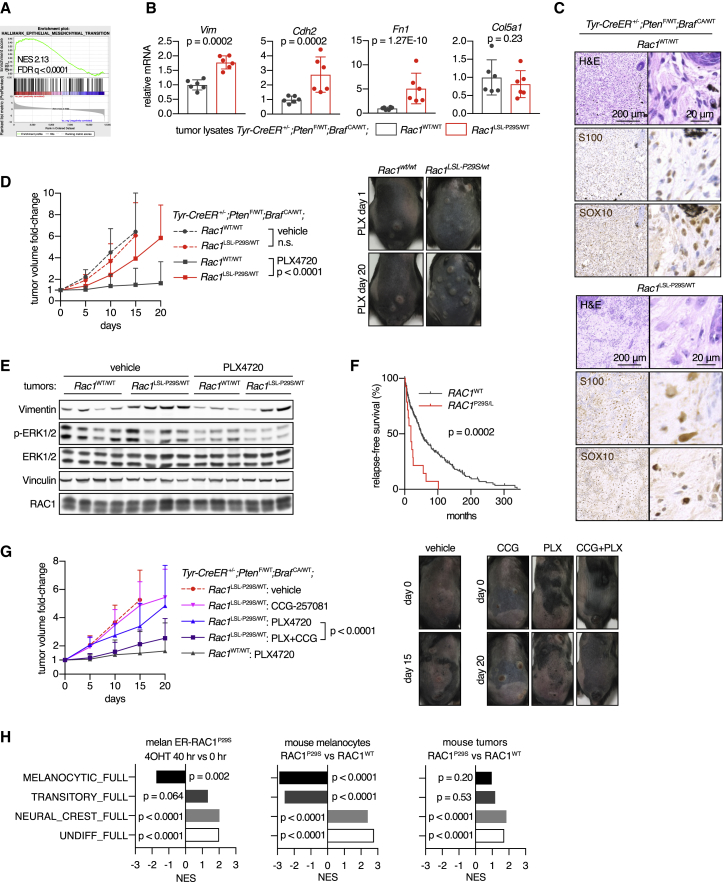
Figure 7.
Effect of RAC1P29S Drug Resistance and Mesenchymal Phenotype in BRAF-Driven Melanoma
(A) GSEA plot of the EMT hallmark gene set from MSigDB, using RNA-seq data of tumor lysates from Tyr-CreER+/–;PtenF/WT;BrafCA/WT;Rac1LSL−P29S/WT mice versus tumor lysates from Tyr-CreER+/–;PtenF/WT;BrafCA/WT;Rac1WT/WT mice (n = 6 tumors from five to six animals per group).
(B) Normalized mRNA read counts of mesenchymal markers in tumor lysates from Tyr-CreER+/–;PtenF/WT;BrafCA/WT mice. For statistical comparison of groups, Wald test was applied in combination with Benjamini-Hochberg correction (n = 6 tumors from five to six animals per genotype).
(C) Effect of expression of RAC1P29S in melanoma on tumor architecture. Representative tumor sections shown stained with H&E or melanoma cell markers S100 and SOX10 (immunohistochemistry).
(D) Tumors were induced in Tyr-CreER+/–;PtenF/WT;BrafCA/WT mice by topical 4OHT. After tumors were established, animals were treated with PLX4720 incorporated in chow. Tumor growth curves were compared using two-way ANOVA, with the p value for the genotype effect indicated (n = 18–44 tumors from 6 to 10 animals per data point). Representative photos of tumors are included.
(E) Tumors at the end of the experiment presented in (D) were harvested, lysed and used for immunoblotting with indicated antibodies.
(F) Influence of RAC1P29S/L mutation status on relapse-free survival in patients with melanoma. Data from the TCGA cutaneous melanoma cohort (n = 309 patients, 45% BRAFmut and 55% BRAFWT) were used. Groups were compared using log rank testing (Mantel-Cox), with the p value indicated.
(G) After tumors were established by topical 4OHT, animals were treated with PLX4720 incorporated in chow and CCG-257081 by oral gavage. Tumor growth curves were compared using two-way ANOVA, with the p value for the genotype effect indicated (n = 10–15 tumors from 3 to 4 animals per data point).
(H) GSEA was performed using the four melanoma differentiation state signatures developed by Tsoi et al. (2018), in combination with RNA-seq dataset of changes in ER-RAC1P29S melanocyte-treated 4OHT (n = 3 independent experiments; left panel), RNA-seq data from melanocytes with endogenous RAC1P29S versus RAC1WT (n = 3 independent cultures per genotype; middle panel) and RNA-seq data of tumor lysates from Tyr-CreER+/–;PtenF/WT;BrafCA/WT;Rac1LSL−P29S/WT mice versus Tyr-CreER+/–;PtenF/WT;BrafCA/WT;Rac1WT/WT mice (n = 6 tumors from five to six animals per group; right panel); p values are indicated.
For all graphs: means ± SD is shown. See also Figure S7.