Figure 4.
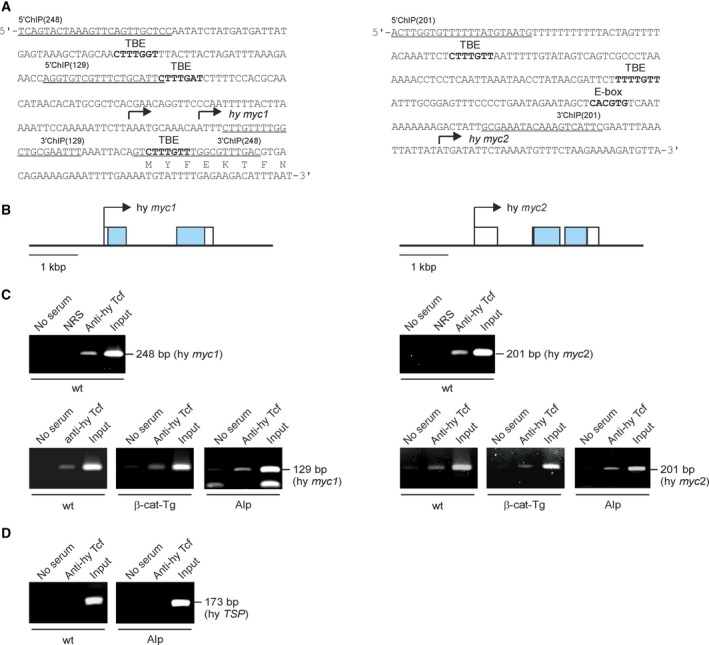
Structures of the Hydra myc1 and myc2 promoter regions and binding of Hydra Tcf. (A) Nucleotide sequences of the myc1 and myc2 regulatory regions. The transcription start sites mapped by 5′RACE (arrows), potential binding sites (in bold) for the transcription factors Tcf (TBE) or Myc (E‐box), and binding sites for 5′ and 3′ ChIP primers (underlined) are indicated. (B) Topographies of Hydra magnipapillata genomic loci (accession nos. NW_004167287, NW_004167363) containing the Hydra myc1 and myc2 genes. Exons are depicted by boxes with the coding regions shown in black. Arrows indicate the main transcription start sites. (C) ChIP of the Hydra myc1 and myc2 promoter regions using chromatin from whole Hydra wild type (wt) animals, from β‐catenin transgenic animals (β‐cat‐Tg), and from Alsterpaullone‐treated (Alp) Hydra. An antiserum directed against Hydra Tcf was used for precipitation, followed by PCR amplification of the indicated fragments from the myc1 or myc2 regulatory regions. Reactions with normal rabbit serum (NRS) or total chromatin (Input) were used as controls. (D) A fragment containing no Tcf binding site derived from the Hydra TSP regulatory region 36 was amplified as a control.
