Figure 1.
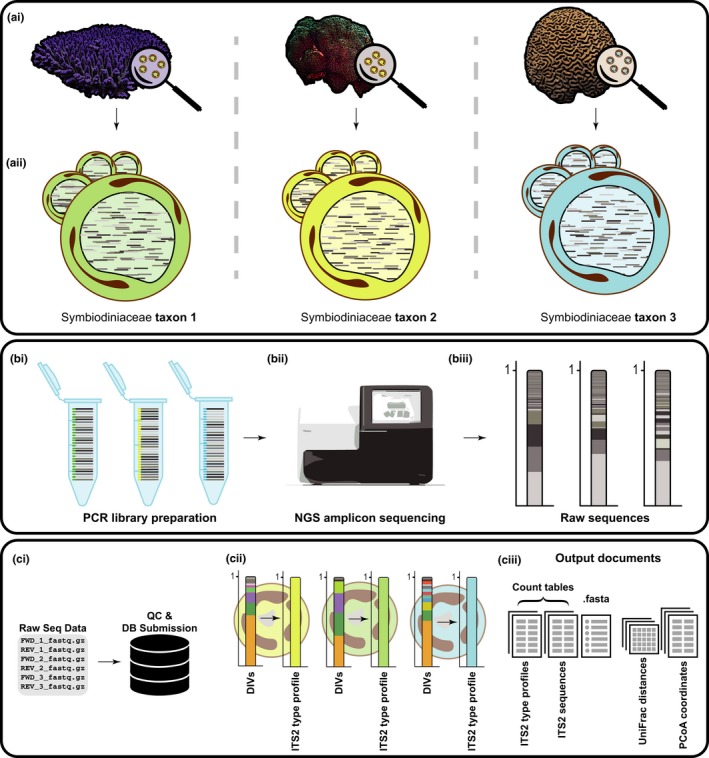
Overview of the typical workflow employing the SymPortal analytical framework. Sampling: (ai) Field sampling: corals containing populations of Symbiodiniaceae are sampled. (aii) Each Symbiodiniaceae cell contains hundreds of copies of the ITS2 gene representing many unique sequences (intragenomic sequence diversity). In this example, three corals are sampled, each containing a single genetically differentiated population of Symbiodiniaceae. PCR library preparation and sequencing: (bi) genomic DNA is extracted from each sample, the ITS2 region is PCR‐amplified and PCR libraries are prepared for sequencing through clean‐up and adapter ligation. (bii) Prepared libraries are sequenced on the Illumina platform producing, (biii) raw sequencing reads in paired.fastq.gz format. SymPortal analysis: (ci) paired.fastq.gz files are quality control processed and loaded into the SymPortal framework's database before being run through the SymPortal analysis. (cii) During the analysis, re‐occurring sets of ITS2 sequences are identified as ‘defining intragenomic [ITS2 sequence] variants’ (DIVs) that are used to define novel or pre‐existing, taxa‐representative, ITS2 type profiles. (ciii) tab‐delimited count tables are output detailing the abundance of predicted ITS2 type profiles and post‐quality control sequences found in each sample. A .fasta is also provided detailing nucleotide sequence information. Finally, clade‐separated UniFrac distance matrices and calculated principal coordinate analysis (PCoA) coordinates are output for between‐sample and between‐ITS2 type profile pairwise comparisons [Colour figure can be viewed at wileyonlinelibrary.com]
