Figure 3.
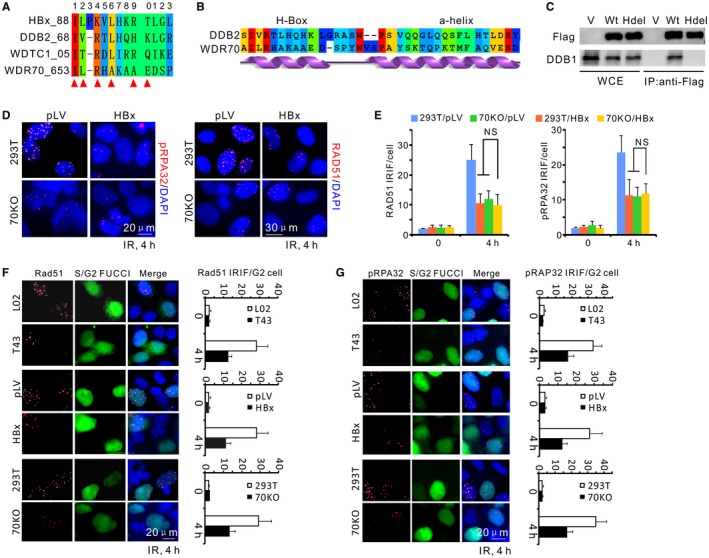
Disruption of CRL4WDR70 complex by HBx results in a resection defect. (A) Sequence alignment for the H‐boxes of cellular DCAF proteins and HBx (WDTC1, alias of DCAF9). Red triangle, binding hotspots serving as key DDB1‐contacting residues. (B) By sequence alignment and secondary structure prediction, the WDR70 C‐terminal sequence possesses a similar H‐box sequence and an adjunct α‐helical structure to that of DDB2, indicating their similar docking mode to DDB1. (C) Immunoprecipitation of endogenous DDB1 in 293T cells by Flag‐tagged WDR70 with or without H‐boxes (amino acids 653‐662). (D) Immunofluorescent staining for phosphorylated RPA32 or RAD51 IRIF in parental and WDR70 knockout 293T cells with or without HBx expression. IRIF defects in 70KO cells were not exacerbated by the presence of HBx. (E) Quantification for IRIF as in (D). n = 3 biological repeats. Error bars = SD. (F,G) Representative images and quantification for RAD51 (F) and pRPA32 (G) foci in IR‐treated cells (4 Gy). Geminin‐green fluorescent protein represents S/G2 cells (S/G2‐FUCCI). Abbreviations: DAPI, 4′,6‐diamidino‐2‐phenylindole; FUCCI, fluorescent ubiquitylation‐based cell cycle indicator; Hdel, H‐box deletion mutant of WDR70; IP, immunoprecipitation; 70KO, WDR70 knockout; LV, lentivirus; NS, no significant difference; WCE, whole cell extracts; V, empty vector.
