Figure 4.
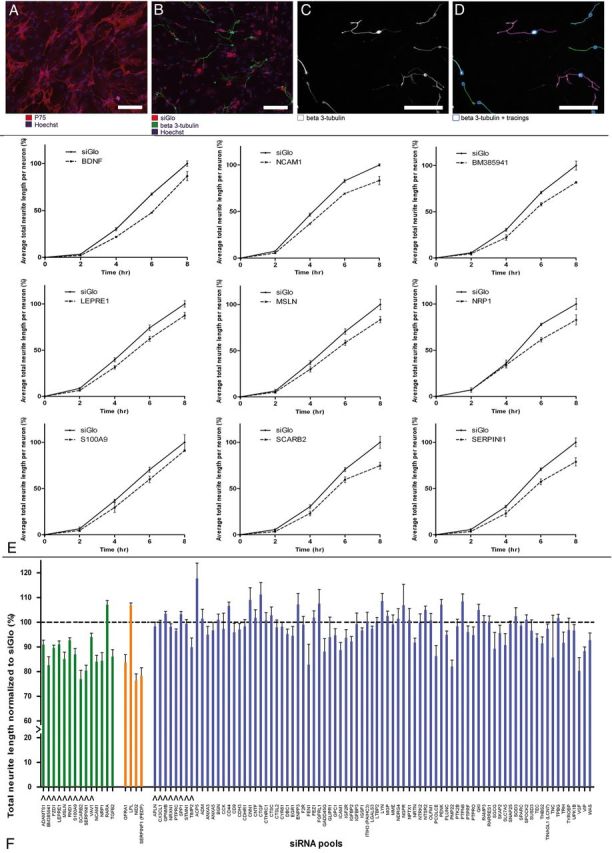
Functional study of candidate genes in siRNA-mediated outgrowth screen. A, OEC feeding layer in a 96-well plate visualized by p75 staining (red) and Hoechst (blue). B, DRGs, labeled for β3-tubulin (green), were plated onto an OEC feeding layer (Hoechst; blue), which was transfected with siGlo (red). C, D, Automated tracings of embryonic DRG cell bodies and neurites, labeled for β3-tubulin (white) by Cellomics technology. Scale bar, 100 μm. E, BDNF and the 8 genes that had a significant effect on neurite outgrowth in siRNA screen Phase 1. Graphs represent total neurite length of embryonic DRGs cultured on OECs transfected with an siRNA pool against BDNF, NCAM1, BM385941, LEPRE1, MSLN, NRP1, S100A9, SCARB2, and SERPIN1. Embryonic DRGs were incubated on the transfected OEC feeding layer for 2, 4, 6, and 8 h. Values are normalized to the siGlo condition. All graphs are the combined result of three independent experiments. F, Graph presenting combined results from siRNA screen Phase 1 and Phase 2. Total neurite length is shown of embryonic DRGs cultured for 8 h on OECs transfected with an siRNA pool against one of all 102 candidate genes. Neurite length was normalized to the siGlo condition (dotted line). Data are presented as the normalized mean ± SEM of the average neurite length. S100A9 was included as an extra gene in Phase 2 to confirm the results obtained for this gene in Phase 1 of the screen. Green and orange bars represent the 18 genes for which knockdown significantly affected neurite length in Phase 1 and 2 of the screen (p < 0.05). Knockdown of 16 genes resulted in a significant reduction of total neurite length, and knockdown of two genes resulted in a significant increase of total neurite length. Orange bars represent genes for which knockdown disrupted the OEC feeding layer. Blue bars represent the 84 genes that had no significant effect on total neurite length. Arrows under the bars indicate the genes that were selected for LV-mediated overexpression experiments.
