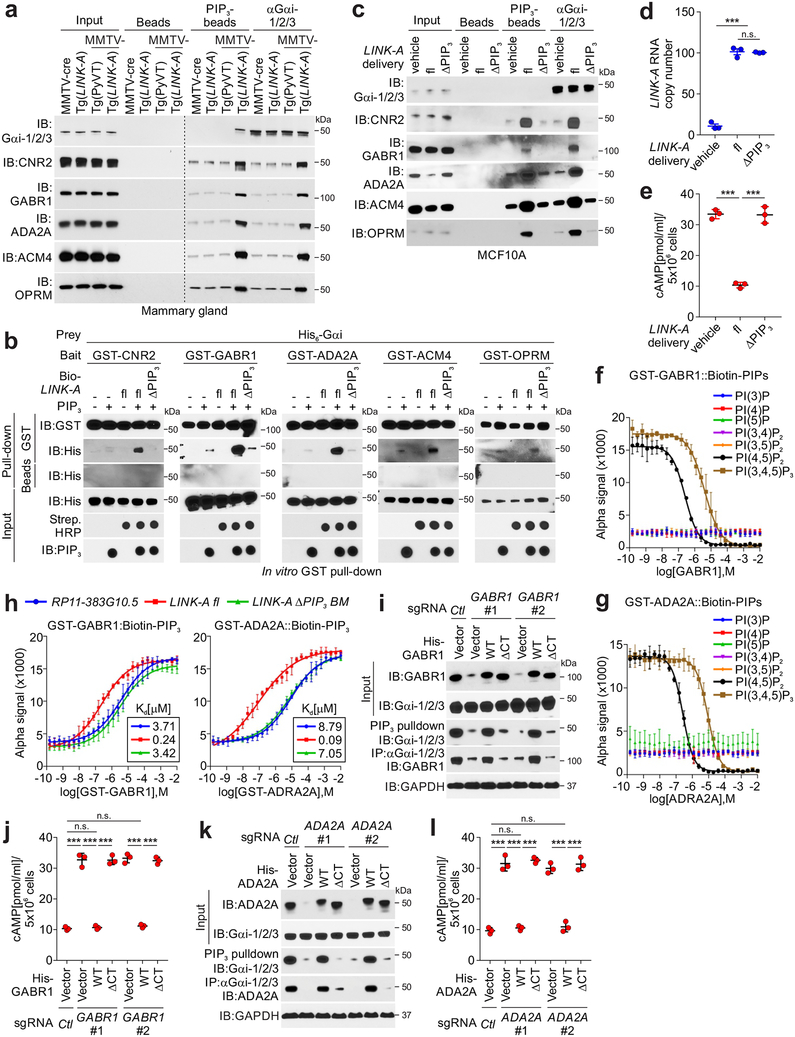
Figure 3: LINK-A mediates crosstalk between PtdIns (3,4,5)P3 and GPCRs.
a, Immunoblotting detection using indicated antibodies of immunoprecipitates using indicated beads or antibodies using MMTV-cre or Tg(LINK-A) normal mammary gland, MMTV-PyVT tumor or MMTV-Tg(LINK-A) tumor lysates. Three independent experiments were performed and yielded similar results. b, GST-pulldown using His-tagged Gαi, GST-tagged GPCRs, and PtdIns (3,4,5)P3, in the presence of biotinylated LINK-A full length (fl) or ΔPtdIns (3,4,5)P3 mutant. Three independent experiments were performed and yielded similar results. c, Immunoblotting detection using indicated antibodies of immunoprecipitates using indicated beads or antibodies of MCF10A cells delivered with vehicle, LINK-A fl or ΔPtdIns (3,4,5)P3 mutant. Three independent experiments were performed and yielded similar results. d and e, Measurement of LINK-A copy number (d), or cellular cAMP level (e) of MCF10A cells delivered with vehicle, LINK-A fl or ΔPtdIns (3,4,5)P3 mutant (n.s., p=0.399, ***p<0.001). Results are mean ± s.e.m. of n=3 independent experiments yielding similar results, P values were determined by one-way ANOVA. f and g, Competition binding assay to determine Kd for interaction between GST-tagged Gαi (left top), or GST-tagged GABR1 (f) or –ADA2A (g) and biotinylated PIPs as indicated. The Kd value (μM) are shown. Results are mean ± s.d. of n=3 independent experiments yielding similar results. h, Competition binding assay to determine Kd for interaction between GST-tagged GPCRs and biotinylated PtdIns (3,4,5)P3 in the presence of RP11–383G10.5, LINK-A fl or LINK-A ΔPtdIns (3,4,5)P3 mutant. Results are mean ± s.d. of n=3 independent experiments. i-l, Immunoblotting detection using indicated antibodies in MDA-MB-231 cells harboring sgRNAs knocking out GABR1 (i, j), ADA2A (l, m) respectively, followed by expression of GPCRs wild type or ΔPtdIns(3,4,5)P3 mutants. The lysates were subjected to immunoblotting detection using indicated antibodies (i, k) or measurement of cAMP concentration (j, l) (n.s., p>0.05, ***p<0.001). Results are mean ± s.d. of n=3 independent experiments, P values were determined by one-way ANOVA.