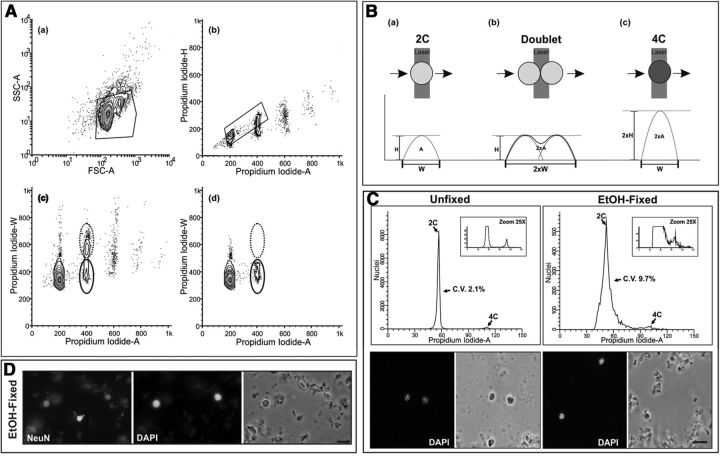
Figure 1.
Electronic gating procedure for murine nuclear samples stained with PI and analyzed by flow cytometry. A, Fresh cell nuclei were isolated from the cerebral cortex of 2-month-old mice, stained with PI, and subjected to flow cytometric analysis. Nuclei were gated (polygonal box) on forward scattering area (FCS-A), a measure of particle size, and side scattering area (SSC-A), a measure of particle complexity (a). DNA content was then assessed from the gated nuclear population by plotting Propidium Iodide-H versus Propidium Iodide-A levels. Diploid and tetraploid nuclei were subsequently gated from these plots (polygonal box), while the doublets of diploid nuclei were discarded (b). DNA content was also assessed from the gated nuclear population in a by plotting Propidium Iodide-W versus Propidium Iodide-A levels. Tetraploid nuclei are surrounded by a solid line while doublets of diploid nuclei are surrounded by a dotted line (c). When DNA content is assessed from the gated nuclear population in b by plotting Propidium Iodide-W versus Propidium Iodide-A levels, doublets of diploid nuclei (dotted line) disappear (d), thus demonstrating that our gating procedure is an efficient method for analysis of diploid and tetraploid (solid line) nuclei. B, Scheme illustrating the method used for doublet discrimination based on the pulse-processing method. When a PI-labeled nucleus passes through the laser beam a fluorescent signal is converted into an electrical pulse defined by its width (W), its maximal intensity (H), and its integrated area (A). The signal recorded from a tetraploid nucleus (c) has a double H value compared with a diploid nucleus (a), whereas a doublet of diploid nuclei (b) results in a fluorescent signal with double W value compared with that of a diploid nucleus. Therefore, tetraploid nuclei can easily be discriminated from doublets of diploid nuclei due to the H/A and the W/A ratio of their PI signal. C, Cell nuclei isolated from the cerebral cortex of 2-month-old mice were either stained with PI and subjected to flow cytometric analysis (Unfixed) or fixed in ice-cold 70% ethanol overnight before staining with PI and flow cytometric analysis (EtOH-fixed). Bottom, Illustrates the aspect of the fresh and ethanol-fixed nuclei, counterstained with DAPI. Top, Illustrates representative DNA content histograms from either unfixed or ethanol-fixed nuclei. Flow cytometric analyses were performed in parallel. CV (C.V.) are shown. Note that the CV is lower when fresh nuclei are analyzed. D, Ethanol-fixed nuclei immunostained with an anti-NeuN antibody and counterstained with DAPI demonstrate a lot of staining background, which likely explains the increased CV. Scale bars: C, 10 μm; D, 20 μm.