Figure 1.
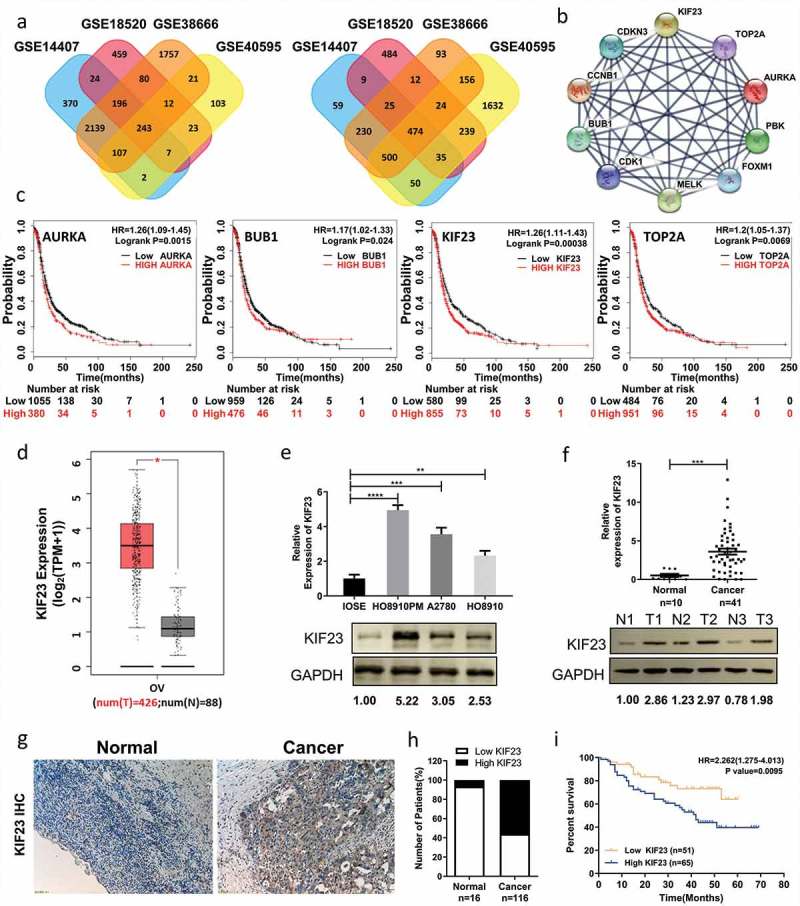
KIF23 expression is significantly up-regulated in ovarian cancer and predicts poor prognosis for ovarian cancer patients.
A: Venn diagrams showed DEGs that had common changes from the four cohort profile data sets (GSE14407, GSE18520, GSE38666 and GSE40595). The DEGs consisting of 243 up-regulated (Left) and 474 down-regulated genes (Right) were detected with p < 0.05 and [logFC] > 1.5 as the cut-off criterion. The overlaps meant the commonly changed DEGs. Different colors were used to distinguish different data sets; B: The protein–protein interaction network of top 10 hub genes; C: Prognostic value of four genes (AURKA, BUB1, KIF23 and TOP2A) in ovarian cancer patients (HR: hazard ratio); D: The expression level of KIF23 in 426 ovarian cancer tissues and 88 normal ovarian tissues from GEPIA (OV: Ovarian serous cystadenocarcinoma; T: Tumor; N: Normal); E: The expression levels of KIF23 mRNA and protein in ovarian epithelial normal cell line and ovarian cancer cell lines; F: The expression levels of KIF23 mRNA and protein in normal ovarian tissues and ovarian cancer tissues (T: Tumor; N: Normal); G: Immunohistochemical analyses of KIF23 in normal ovarian tissues (n = 16) and ovarian cancer tissues (n = 116) (x100); Expression of KIF23 was significantly up-regulated in ovarian cancer tissues; H: Bar plot showed the proportion of high or low expression of KIF23 in normal or ovarian cancer tissues; I: Overall survival analysis of KIF23 expression revealed that high expression of KIF23 is associated with poor prognosis in 116 ovarian cancer patients (p = 0.0095); Blue and orange represented the high and low expression of KIF23, respectively. (Data were shown by mean ± SEM from three independent experiments; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, as compared to date from control groups).
