Figure 2.
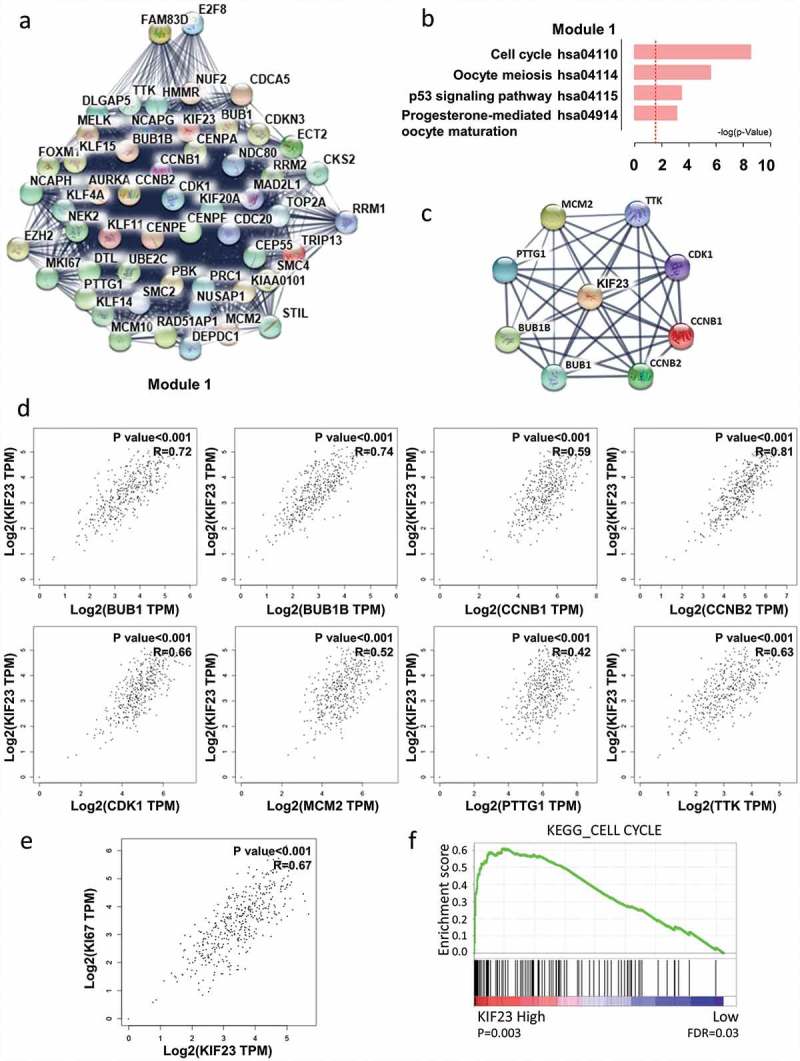
The correlation analyses between KIF23 and cell cycle-related proteins.
A: Using the STRING online database, total of 717 DEGs were filtered into the DEGs protein–protein interaction (PPI) network complex. PPI network modules were screened by Molecular Complex Detection (MCODE) plug-in. The most significantly functional module in the protein–protein interaction network was showed; B: Enrichment of signaling pathways of DEGs conducted in the module; C: The PPI network of KIF23 and cell cycle-related genes (BUB1, BUB1B, CCNB1, CCNB2, CDK1, MCM2, PTTG1 and TTK); D: The correlation analyses from GEPIA between KIF23 and cell cycle-related genes. Significant correlations were observed between KIF23 mRNA expression and mRNA expression of BUB1 (R = 0.72, P < 0.001), BUB1B (R = 0.74, P < 0.001), CCNB1 (R = 0.59, P < 0.001), CCNB2 (R = 0.81, P < 0.001), CDK1 (R = 0.66, P < 0.001), MCM2 (R = 0.52, P < 0.001), PTTG1 (R = 0.42, P < 0.001), TTK (R = 0.63, P < 0.001) (R: Pearson Correlation Coefficient); E: The expression of KIF23 mRNA is positively correlated with the expression of Ki67 mRNA analysed by GEPIA in ovarian serous cystadenocarcinoma tissues from TCGA (R = 0.67, P < 0.001); F: GSEA indicating KEGG CELL CYCLE signatures pathways was more correlated with patients with KIF23 high versus KIF23 low patients (FDR: False Discovery Rate).
