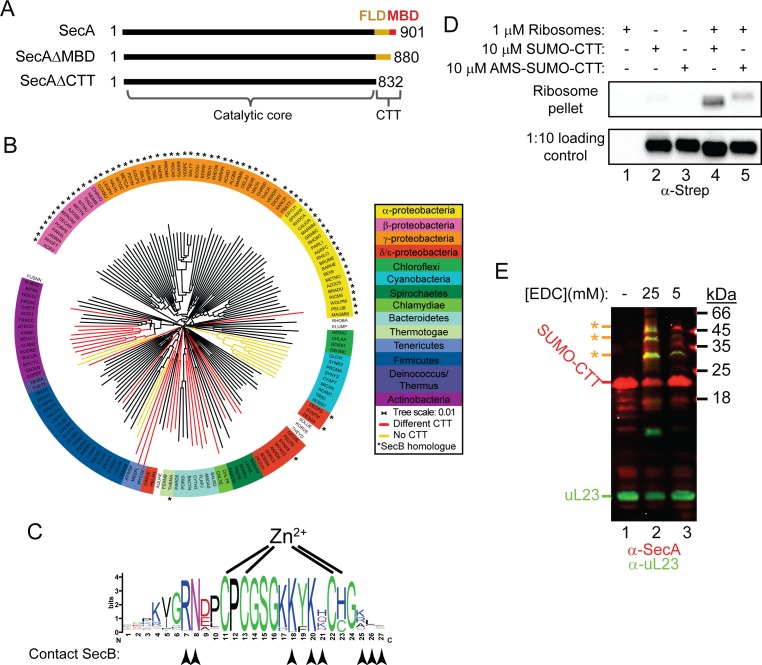
Figure 1. Phylogenetic analysis of the CTT and binding of E. coli CTT to the ribosome.
(A) Schematic diagram of the primary structure of SecA, SecAΔMBD and SecAΔCTT. Structures are oriented with the N-termini to the left, and the amino acid positions of the N- and C-termini are indicated. Residues of the catalytic core and the CTT are indicated below. Catalytic core, black; FLD, yellow; MBD, red. (B) Phylogenetic tree of the SecA proteins of 156 representative species from 155 different bacterial families. Species names are given as the five-letter organism mnemonic in UniProtKB (The UniProt Consortium, 2017). Taxonimical classes are colour-coded according to the legend. Leaves representing SecA proteins with an MBD are coloured black. Those with CTTs lacking a MBD are coloured red, and those that lack a CTT entirely are coloured yellow. Species that also contain a SecB protein are indicated with a star (*). (C) Logo of the consensus sequence of the MBD generated from the 117 species containing the MBD in the phylogenetic analysis. Positions of the metal-coordinating amino acids are indicated above. Amino acids that contact SecB in the structure of the MBD-SecB complex (Zhou and Xu, 2003) (1OZB) are indicated by arrowheads below. (D) Binding reactions containing 1 μM ribosomes, 10 μM SUMO-CTT and 10 μM AMS-modified SUMO-CTT (AMS-SUMO-CTT) were equilibrated at room temperature and layered on a 30% sucrose cushion. Ribosomes were then sedimented through the cushion by ultracentrifugation. Samples were resolved on SDS-PAGE and probed by western blotting against the Strep tag using HRP-coupled Streptactin. (E) 10 μM SUMO-CTT containing an N-terminal Strep(II)-tag was incubated with 1 μM purified ribosomes and treated with 5 mM or 25 mM EDC, as indicated. Samples were resolved by SDS-PAGE and analysed by western blotting by simultaneously probing against SecA (red) and ribosomal protein uL23 (green). The positions of SUMO-CTT, L23 and crosslinking adducts between them (*) are indicated at left.
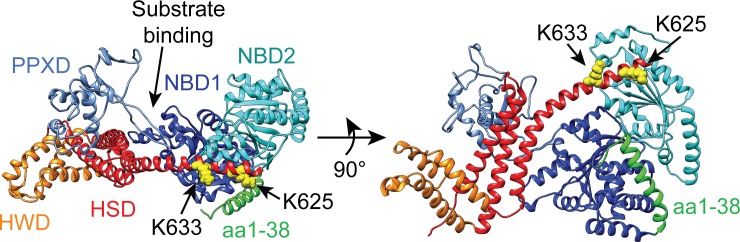
Figure 1—figure supplement 1. Structural model of the catalytic core of SecA in the ‘closed’ conformation.
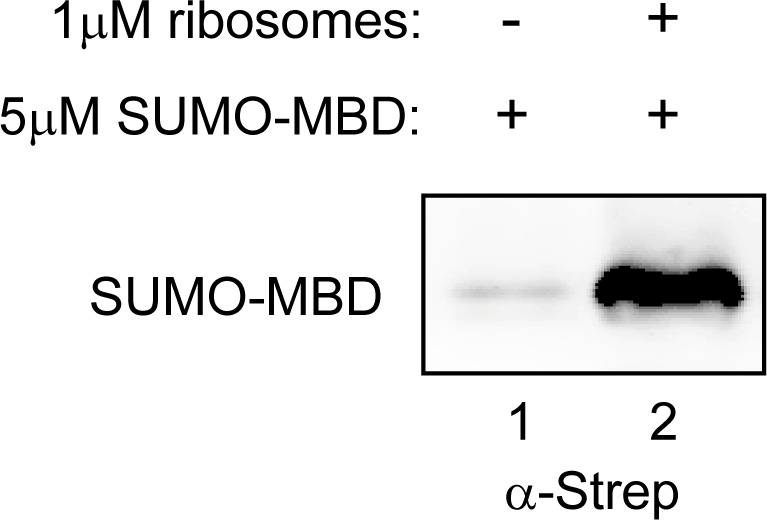
Figure 1—figure supplement 2. SUMO-MBD cosediments with ribosomes.